Journal:MicroPubl Biol:000606
From Proteopedia
(Difference between revisions)

| Line 8: | Line 8: | ||
Normally, changes in a protein sequence resulting in a gain or loss of function are constrained by selective pressures. However, when a gene duplication event occurs, one of the two gene products can continue to fulfill the original function(s). The second gene product is then under reduced selective pressures and is free to accumulate sequence changes that can result in the loss of some of the original function(s). These sequence changes, such as the loss of catalytic residues, can result in the formation of a pseudoenzyme, a protein that is catalytically inactive but homologous to a functional, catalytic enzyme family <ref name='MurphyFarhanEyers2017'>PMID: 28408493</ref>. This was first seen in catalytically dead kinases - the so-called pseudokinases - where it is estimated that 10%, or 50 out of 500, human kinases are pseudokinases. Pseudoenzymes as functional but catalytically dead enzymes are in contrast to pseudogenes. Both share the property of possessing sequence changes from their ancestral counterpart. However, pseudogenes are typically non-coding such as through the loss of start codons and sequence frameshifts. Based on previous work <ref name='CharetteBaserga2010'/>, we propose that a DEAD-box RNA helicase underwent a gene duplication in the last common eukaryotic ancestor. RNA helicases possess two functions - RNA binding and ATP binding/hydrolysis. Thus, we suggest that the catalytic residues responsible for ATP binding and hydrolysis were lost - hence being a pseudohelicase - while the RNA binding function was retained in what might be a case of evolutionary subfunctionalization. (It remains to be determined if Utp25s RNA binding activity <ref name='CharetteBaserga2010'/><ref name='GoldfederOliveira2010'/>) is direct or mediated through an unknown RNA-binding protein). The continued cataloging of pseudoenzymes, such as this identification of pseudohelicases as a new category, will increase our understanding of pseudoenzyme function and protein evolution. As pseudoenzymes participate in signal transduction (as allosteric regulators and pseudokinases) and ribosome assembly (here as pseudohelicases), they expand our knowledge of cell mechanisms and are potential new drug targets for diseases such as cancer. | Normally, changes in a protein sequence resulting in a gain or loss of function are constrained by selective pressures. However, when a gene duplication event occurs, one of the two gene products can continue to fulfill the original function(s). The second gene product is then under reduced selective pressures and is free to accumulate sequence changes that can result in the loss of some of the original function(s). These sequence changes, such as the loss of catalytic residues, can result in the formation of a pseudoenzyme, a protein that is catalytically inactive but homologous to a functional, catalytic enzyme family <ref name='MurphyFarhanEyers2017'>PMID: 28408493</ref>. This was first seen in catalytically dead kinases - the so-called pseudokinases - where it is estimated that 10%, or 50 out of 500, human kinases are pseudokinases. Pseudoenzymes as functional but catalytically dead enzymes are in contrast to pseudogenes. Both share the property of possessing sequence changes from their ancestral counterpart. However, pseudogenes are typically non-coding such as through the loss of start codons and sequence frameshifts. Based on previous work <ref name='CharetteBaserga2010'/>, we propose that a DEAD-box RNA helicase underwent a gene duplication in the last common eukaryotic ancestor. RNA helicases possess two functions - RNA binding and ATP binding/hydrolysis. Thus, we suggest that the catalytic residues responsible for ATP binding and hydrolysis were lost - hence being a pseudohelicase - while the RNA binding function was retained in what might be a case of evolutionary subfunctionalization. (It remains to be determined if Utp25s RNA binding activity <ref name='CharetteBaserga2010'/><ref name='GoldfederOliveira2010'/>) is direct or mediated through an unknown RNA-binding protein). The continued cataloging of pseudoenzymes, such as this identification of pseudohelicases as a new category, will increase our understanding of pseudoenzyme function and protein evolution. As pseudoenzymes participate in signal transduction (as allosteric regulators and pseudokinases) and ribosome assembly (here as pseudohelicases), they expand our knowledge of cell mechanisms and are potential new drug targets for diseases such as cancer. | ||
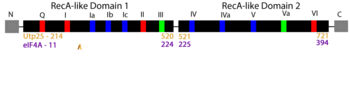
| - | RNA helicases comprise a large family of proteins involved in nearly all aspects of RNA function, including ribosome assembly and translation. Historically, the main ascribed function of helicases is to unwind (''i.e.'' unzip) DNA and RNA duplexes, though their function is now recognized to include the association, dissociation, or remodeling of RNA-RNA and RNA-protein complexes. The focus of our article, Utp25, was first discovered as a component of the small subunit (SSU) processome, a 6 MDa ribonucleoprotein complex responsible for most pre-rRNA processing and assembly steps of the SSU of the ribosome <ref name='CharetteBaserga2010'/><ref name='GoldfederOliveira2010'/>). Sequence analysis identified in Utp25 partial sequence motifs | + | RNA helicases comprise a large family of proteins involved in nearly all aspects of RNA function, including ribosome assembly and translation. Historically, the main ascribed function of helicases is to unwind (''i.e.'' unzip) DNA and RNA duplexes, though their function is now recognized to include the association, dissociation, or remodeling of RNA-RNA and RNA-protein complexes. The focus of our article, Utp25, was first discovered as a component of the small subunit (SSU) processome, a 6 MDa ribonucleoprotein complex responsible for most pre-rRNA processing and assembly steps of the SSU of the ribosome <ref name='CharetteBaserga2010'/><ref name='GoldfederOliveira2010'/>). Sequence analysis identified in Utp25 partial sequence motifs that are hallmarks of the DEAD-box family of RNA helicases. |
| + | [[Image:AlignFig1r.png|left|350px|thumb|Linear arrangement of the DEAD-box family motifs, with sequences mediating RNA binding in blue, ATP binding and hydrolysis in red, and those linking ATP and RNA binding in green (adapted from Fig 1a; (Putnam and Jankowsky 2013))]] | ||
| + | {{Clear}} | ||
| + | |||
| + | These conserved motifs mediate the binding of RNA and/or the binding and hydrolysis of ATP. Interestingly, Utp25 has significant sequence changes in most of these motifs. Mutational loss of the remaining conserved sequence motifs 1a and partial motif VI resulted in no change in growth <ref name='CharetteBaserga2010'/>. | ||
Using the AlphaFold predicted yeast Utp25 structure as a query, we used Dali to search for proteins with a similar structure. Our top hits were to other DEAD-box helicases including the prototypical RNA helicase eIF4A. We then used Chimera to structurally align Utp25 and eIF4a. By independently aligning the structures of <scene name='92/920755/Cv/9'>domain 1</scene> from Utp25 (gold) and eIF4A (medium violet red) and similarly of <scene name='92/920755/Cv4/2'>domain 2</scene>, we show that Utp25 globally adopts a structure that is very similar to that of DEAD-box RNA helicases. | Using the AlphaFold predicted yeast Utp25 structure as a query, we used Dali to search for proteins with a similar structure. Our top hits were to other DEAD-box helicases including the prototypical RNA helicase eIF4A. We then used Chimera to structurally align Utp25 and eIF4a. By independently aligning the structures of <scene name='92/920755/Cv/9'>domain 1</scene> from Utp25 (gold) and eIF4A (medium violet red) and similarly of <scene name='92/920755/Cv4/2'>domain 2</scene>, we show that Utp25 globally adopts a structure that is very similar to that of DEAD-box RNA helicases. | ||
Revision as of 14:24, 4 September 2022
| |||||||||||
This page complements a publication in scientific journals and is one of the Proteopedia's Interactive 3D Complement pages. For aditional details please see I3DC.