We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1732
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
== Structural highlights == | == Structural highlights == | ||
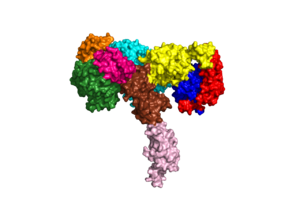
An insulin receptor is a dimer of heterodimers. The dimers are noncovalent, but the insulin receptors are covalently maintained as functional dimers by disulfide bonds. An insulin receptor is comprised of 2 α-chains, and 2 β-chains. The α-chain and an estimated 190 residues of the β-chain are located on the extracellular side of the plasma membrane. The rest of the beta-chain consists of a single transmembrane helix, the juxtamembrane domain, and the intracellular tyrosine kinase domain. | An insulin receptor is a dimer of heterodimers. The dimers are noncovalent, but the insulin receptors are covalently maintained as functional dimers by disulfide bonds. An insulin receptor is comprised of 2 α-chains, and 2 β-chains. The α-chain and an estimated 190 residues of the β-chain are located on the extracellular side of the plasma membrane. The rest of the beta-chain consists of a single transmembrane helix, the juxtamembrane domain, and the intracellular tyrosine kinase domain. | ||
| - | The | + | The alpha subunits are the site for insulin binding. Each subunit is comprised of 2 Leucine rich domains (L1 and L2), a Cysteine rich domain (CR) and an α-chain C-terminal helix (α-CT). The two subunits are held together by a disulfide bond between the cysteine rich domains. |
| - | + | [[Image:6CE7.png]] | |
| + | Due to the heterodimeric nature of the receptor, there are two types of insulin binding sites that are split into pairs in the alpha subunits: sites 1 and 1' and sites 2 and 2', for a total of 4 binding sites of insulin. Binding sites 1 and 1' have a greater surface area and are more easily accessible for the insulin to bond, resulting in a higher affinity for insulin binding. Binding sites 2 and 2' have less surface area and are located on the back of the beta sheet so their binding sites do not get filled as quickly. | ||
| + | |||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 18:28, 14 November 2022
| This Sandbox is Reserved from August 30, 2022 through May 31, 2023 for use in the course Biochemistry I taught by Kimberly Lane at the Radford University, Radford, VA, USA. This reservation includes Sandbox Reserved 1730 through Sandbox Reserved 1749. |
To get started:
More help: Help:Editing |
Your Heading Here (maybe something like 'Structure')
| |||||||||||
References
1. PDB101: Molecule of the month: Insulin receptor https://pdb101.rcsb.org/motm/182 (accessed Oct 21, 2022).