We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1732
From Proteopedia
(Difference between revisions)
| Line 25: | Line 25: | ||
[[Image:6CE7.png]] | [[Image:6CE7.png]] | ||
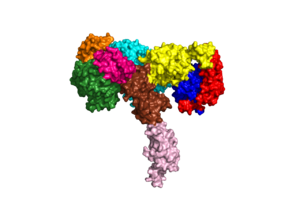
| - | Due to the heterodimeric nature of the receptor, there are two types of insulin binding sites that are split into pairs in the alpha subunits: sites 1 and 1' and sites 2 and 2', for a total of 4 binding sites of insulin. Binding sites 1 and 1' have a greater surface area and are more easily accessible for the insulin to bond, resulting in a higher affinity for insulin binding. Binding sites 2 and 2' have less surface area and are located on the back of the beta sheet so their binding sites do not get filled as quickly. | + | Due to the heterodimeric nature of the receptor, there are two types of insulin binding sites that are split into pairs in the alpha subunits: sites 1 and 1' and sites 2 and 2', for a total of 4 binding sites of insulin. Binding sites 1 and 1' have a greater surface area and are more easily accessible for the insulin to bond, resulting in a higher affinity for insulin binding. Binding sites 2 and 2' have less surface area and are located on the back of the beta sheet so their binding sites do not get filled as quickly.The beta subunits consist of Fibronectin domain III-2 (FnIII-2) and Fibronectin domain III-3 (FnIII-3). |
[[Image:biochemIBSpt2.png]] | [[Image:biochemIBSpt2.png]] | ||
| + | |||
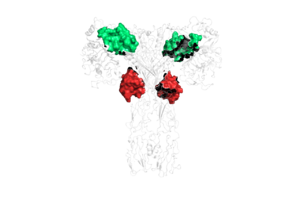
| + | There are two main conformations of full-length insulin receptors, U-shaped and T-shaped. The U-shaped conformation exists when there is an absence of insulin on the protein, whereas the T-shaped and II-shaped conformations exist in the presence of insulin. The U-shaped conformation is also known as the auto-inhibited state. In this state, the L1 domain contacts the L1’ and L2’ domains of the partnering protein. When insulin binds to the receptor protein, it causes a conformational change from the U-shape to the T-shape conformation. | ||
This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene> by Group, and another to make <scene name="/12/3456/Sample/2">a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
Revision as of 23:30, 14 November 2022
| This Sandbox is Reserved from August 30, 2022 through May 31, 2023 for use in the course Biochemistry I taught by Kimberly Lane at the Radford University, Radford, VA, USA. This reservation includes Sandbox Reserved 1730 through Sandbox Reserved 1749. |
To get started:
More help: Help:Editing |
Insulin Receptor Protein
| |||||||||||
References
1. PDB101: Molecule of the month: Insulin receptor https://pdb101.rcsb.org/motm/182 (accessed Oct 21, 2022).