Androgen receptor
From Proteopedia
(Difference between revisions)
| Line 9: | Line 9: | ||
===N-terminal Domain (NTD)(residues 1-555)=== | ===N-terminal Domain (NTD)(residues 1-555)=== | ||
| - | This region is required for full transcriptional activity <ref name="Structure">PMID: 24909511</ref> | + | This region is required for full transcriptional activity <ref name="Structure">PMID: 24909511</ref> because of its necessary presence for Ligand-Binding Domain (LBD) activation <ref name="AR">PMID: 33076388</ref>. The sequence and lengths of the polyglutamine (CAG) and polyglycine (GGC) repeats are highly variable in the human population, making this the most variable domain. It has been shown that the length of the CAG repeat region affects folding and structure of this domain. Shorter repeats generally impose a higher AR transactivation activity, whereas longer repeats cause reduced activity <ref name="Structure" />. In healthy people, one region of the AR gene shows up to 36 repeats of the CAG sequence. Patients with abnormally high numbers of CAG repeats can develop spinal muscular atrophy. |
===DNA-Binding Domain (DBD) (residues 555-623)=== | ===DNA-Binding Domain (DBD) (residues 555-623)=== | ||
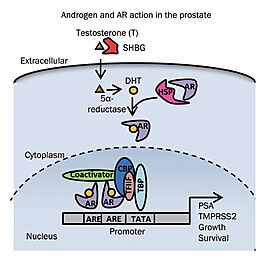
| - | DBD is a cysteine-rich region that is the most highly conserved in the steroid hormone nuclear receptor family <ref name="Structure" /> | + | DBD is a cysteine-rich region that is the most highly conserved one in the steroid hormone nuclear receptor family <ref name="Structure" />. It has been shown that binding of this region to selective androgen response elements (AREs) allow the specific activation functions of the AR. AREs facilitate direct DNA binding of the AR to the promoter and enhancer regions of AR-regulated genes, thereby allowing the activation functions of the N-terminal and LBD to stimulate or repress transcription of these genes <ref name="Bench to Bedside" />. |
| - | AR | + | AR dimeric structure, like that of other steroid receptors, consists of two equal, common hexameric half-sites, separated by a 3 base-pair spacer <ref name="Structure" />. This domain is critical for AR function, due to its role in dimerization and binding of dimerized AR to select motifs on target DNA <ref name="AR" />. |
| - | Each DBD monomer has a core composed of two zinc finger motifs, which | + | Each DBD monomer has a core composed of two zinc finger motifs, which consist of four cysteine residues that coordinate a zinc ion <ref name="Structure" />. The first one is closer to the NTD which has the P box, identical in all family members, that controls the DNA binding specificity at AREs <ref name="AR" />. The second zinc finger motif facilitates AR dimerization via the D box. |
| - | Additionally, a nuclear localisation signal (NLS) | + | Additionally, a nuclear localisation signal (NLS) lies at the junction between the DBD and the hinge region. This NLS binds to importin-α and facilitates nuclear translocation <ref name="AR" />. Passive transport across the nuclear pore complex has been suggested ranging from 20–40 kDa; in contrast, the AR, which is 110 kDa in size, requires importins to be actively transported upon ligand binding <ref name="Structure" />. |
| - | The DBD is linked to the LBD by a flexible hinge region (residues 623-665), which is a | + | The DBD is linked to the LBD by a flexible hinge region (residues 623-665), which is a poorly conserved linker. Once in the nucleus, this region also interacts with the DBD to identify specific sequences for AR binding. It controls the AR activation and degradation. Consequently, mutations in the hinge region can lead to enhanced AR potency <ref name="AR" />. |
===Ligand-Binding Domain (LBD) (residues 665-919)=== | ===Ligand-Binding Domain (LBD) (residues 665-919)=== | ||
| - | The LBD, located at the C- | + | The LBD, located at the C-terminus, is the main target of AR inhibitors <ref name="AR" />. It consists of eleven α-helixes in the ligand binding pocket ( <scene name='54/543362/Cv/3'>active site</scene> ) , wich reposition upon androgen binding, converting this domain into an activation function 2 (AF-2) domain. Unlike other nuclear receptors, the AR does not have H2, which is instead replaced by a long flexible linker <ref name="Structure" />. The LBD binds to motifs in the NTD and in AR-specific cofactors and coactivators. Moreover, LBD-LBD homodimerization of AR is essential for the proper functioning of the receptors <ref name="AR" />. |
| - | This domain has been structurally well characterized by crystallography and a number of mutations have been identified. It is important because not all mutations affect ligand binding, but some of them may disrupt androgen induced | + | This domain has been structurally well characterized by crystallography and a number of mutations have been identified. It is important because not all mutations affect ligand binding, but some of them may disrupt androgen-induced interactions of the N-terminal motif and C-terminal AF-2 <ref name="AR" />. |
==Transcriptional Activation Function== | ==Transcriptional Activation Function== | ||
Two transcriptional activation functions have been identified: | Two transcriptional activation functions have been identified: | ||
| - | *The ligand-independent AF-1 (residues 142-485): located in the NTD is constitutively active. It is the main region responsible for mediating AR transcription. Its contains two | + | *The ligand-independent AF-1 domain (residues 142-485): located in the NTD is constitutively active. It is the main region responsible for mediating AR transcription. Its contains two independent transcription activation units that are indispensable for full activity of the AR <ref name="Structure" />. |
*The ligand-dependent AF-2: is located in the LBD <ref name="Bench to Bedside" />. <scene name='54/543362/H12_androgen_receptor/1'>Helix 12 (H12)</scene> forms the core of this region and acts as a lid to close the LBD upon agonist binding <ref name="Structure" />. It is important for forming the coregulator bindings site as well as mediating direct interactions between the N-terminal and LBD. Key differences in the contribution of specific conserved residues in the AF-2 core domain between the AR and other steroid hormone nuclear receptors have been identified, it would explain the differences in the structure and the function, as well as the coregulatory proteins they interact with <ref name="Bench to Bedside" />. | *The ligand-dependent AF-2: is located in the LBD <ref name="Bench to Bedside" />. <scene name='54/543362/H12_androgen_receptor/1'>Helix 12 (H12)</scene> forms the core of this region and acts as a lid to close the LBD upon agonist binding <ref name="Structure" />. It is important for forming the coregulator bindings site as well as mediating direct interactions between the N-terminal and LBD. Key differences in the contribution of specific conserved residues in the AF-2 core domain between the AR and other steroid hormone nuclear receptors have been identified, it would explain the differences in the structure and the function, as well as the coregulatory proteins they interact with <ref name="Bench to Bedside" />. | ||
| Line 46: | Line 46: | ||
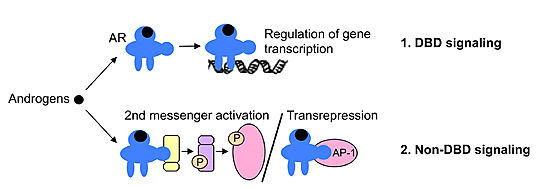
It has been shown that the androgen/AR complex activates 2nd messenger pathways including ERK, Akt and MAPK, and it also interferes with several key proteins including forkhead box protein A1 (FOXA1), PI3K and receptor tyrosine kinases. These effects occur within seconds to minutes of androgen treatment <ref name="Bench to Bedside" /><ref>PMID: 28301631</ref>. | It has been shown that the androgen/AR complex activates 2nd messenger pathways including ERK, Akt and MAPK, and it also interferes with several key proteins including forkhead box protein A1 (FOXA1), PI3K and receptor tyrosine kinases. These effects occur within seconds to minutes of androgen treatment <ref name="Bench to Bedside" /><ref>PMID: 28301631</ref>. | ||
| - | + | Several studies suggest that some of the non-DNA binding-dependent actions of androgens are mediated via the activation of membrane-bound protein receptors. For instance, the iron-regulated transporter-like protein 9 (ZIP9) mediates the androgen-induced apoptosis of ovarian follicle cells, prostate and breast cancer cells <ref name="Bench to Bedside" />. | |
Although the physiological significance of the non-DNA binding-dependent actions of the AR is not yet fully defined, it has been proposed they may oppose the DNA binding-dependent actions and serve as a brake to fine-tune androgen action in target tissues <ref name="Bench to Bedside" />. | Although the physiological significance of the non-DNA binding-dependent actions of the AR is not yet fully defined, it has been proposed they may oppose the DNA binding-dependent actions and serve as a brake to fine-tune androgen action in target tissues <ref name="Bench to Bedside" />. | ||
Revision as of 09:16, 29 November 2022
| |||||||||||
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 Davey RA, Grossmann M. Androgen Receptor Structure, Function and Biology: From Bench to Bedside. Clin Biochem Rev. 2016 Feb;37(1):3-15. PMID:27057074
- ↑ Kolyvas EA, Caldas C, Kelly K, Ahmad SS. Androgen receptor function and targeted therapeutics across breast cancer subtypes. Breast Cancer Res. 2022 Nov 14;24(1):79. doi: 10.1186/s13058-022-01574-4. PMID:36376977 doi:http://dx.doi.org/10.1186/s13058-022-01574-4
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015 Jan;36(1):3-23. doi: 10.1038/aps.2014.18. Epub 2014 Jun , 9. PMID:24909511 doi:http://dx.doi.org/10.1038/aps.2014.18
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 Messner EA, Steele TM, Tsamouri MM, Hejazi N, Gao AC, Mudryj M, Ghosh PM. The Androgen Receptor in Prostate Cancer: Effect of Structure, Ligands and Spliced Variants on Therapy. Biomedicines. 2020 Oct 15;8(10):422. doi: 10.3390/biomedicines8100422. PMID:33076388 doi:http://dx.doi.org/10.3390/biomedicines8100422
- ↑ van Royen ME, van Cappellen WA, de Vos C, Houtsmuller AB, Trapman J. Stepwise androgen receptor dimerization. J Cell Sci. 2012 Apr 15;125(Pt 8):1970-9. doi: 10.1242/jcs.096792. Epub 2012 Feb , 10. PMID:22328501 doi:http://dx.doi.org/10.1242/jcs.096792
- ↑ Kono M, Fujii T, Lim B, Karuturi MS, Tripathy D, Ueno NT. Androgen Receptor Function and Androgen Receptor-Targeted Therapies in Breast Cancer: A Review. JAMA Oncol. 2017 Sep 1;3(9):1266-1273. doi: 10.1001/jamaoncol.2016.4975. PMID:28301631 doi:http://dx.doi.org/10.1001/jamaoncol.2016.4975
- ↑ 7.0 7.1 7.2 Gibson DA, Saunders PTK, McEwan IJ. Androgens and androgen receptor: Above and beyond. Mol Cell Endocrinol. 2018 Apr 15;465:1-3. doi: 10.1016/j.mce.2018.02.013. Epub , 2018 Feb 24. PMID:29481861 doi:http://dx.doi.org/10.1016/j.mce.2018.02.013
- ↑ Ceruti JM, Leiros GJ, Balana ME. Androgens and androgen receptor action in skin and hair follicles. Mol Cell Endocrinol. 2018 Apr 15;465:122-133. doi: 10.1016/j.mce.2017.09.009. , Epub 2017 Sep 12. PMID:28912032 doi:http://dx.doi.org/10.1016/j.mce.2017.09.009
- ↑ 9.0 9.1 9.2 9.3 9.4 9.5 9.6 Solomon ZJ, Mirabal JR, Mazur DJ, Kohn TP, Lipshultz LI, Pastuszak AW. Selective Androgen Receptor Modulators: Current Knowledge and Clinical Applications. Sex Med Rev. 2019 Jan;7(1):84-94. doi: 10.1016/j.sxmr.2018.09.006. Epub 2018 Nov , 30. PMID:30503797 doi:http://dx.doi.org/10.1016/j.sxmr.2018.09.006
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 10.7 Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, Lundasen T, Hughes T, Kojetin DJ. Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev. 2013 Mar 1;65(2):710-78. doi: 10.1124/pr.112.006833. Print 2013 , Apr. PMID:23457206 doi:http://dx.doi.org/10.1124/pr.112.006833
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 11.6 11.7 Christiansen AR, Lipshultz LI, Hotaling JM, Pastuszak AW. Selective androgen receptor modulators: the future of androgen therapy? Transl Androl Urol. 2020 Mar;9(Suppl 2):S135-S148. doi: 10.21037/tau.2019.11.02. PMID:32257854 doi:http://dx.doi.org/10.21037/tau.2019.11.02
- ↑ 12.0 12.1 12.2 12.3 Narayanan R, Coss CC, Dalton JT. Development of selective androgen receptor modulators (SARMs). Mol Cell Endocrinol. 2018 Apr 15;465:134-142. doi: 10.1016/j.mce.2017.06.013. , Epub 2017 Jun 15. PMID:28624515 doi:http://dx.doi.org/10.1016/j.mce.2017.06.013
- ↑ Culig Z, Klocker H, Bartsch G, Hobisch A. Androgen receptors in prostate cancer. Endocr Relat Cancer. 2002 Sep;9(3):155-70. doi: 10.1677/erc.0.0090155. PMID:12237244 doi:http://dx.doi.org/10.1677/erc.0.0090155
- ↑ 14.00 14.01 14.02 14.03 14.04 14.05 14.06 14.07 14.08 14.09 14.10 14.11 14.12 14.13 14.14 14.15 14.16 14.17 14.18 Helsen C, Van den Broeck T, Voet A, Prekovic S, Van Poppel H, Joniau S, Claessens F. Androgen receptor antagonists for prostate cancer therapy. Endocr Relat Cancer. 2014 Aug;21(4):T105-18. doi: 10.1530/ERC-13-0545. Epub 2014 , Mar 17. PMID:24639562 doi:http://dx.doi.org/10.1530/ERC-13-0545
- ↑ 15.0 15.1 15.2 15.3 15.4 15.5 Sesti F, Pofi R, Minnetti M, Tenuta M, Gianfrilli D, Isidori AM. Late-onset hypogonadism: Reductio ad absurdum of the cardiovascular risk-benefit of testosterone replacement therapy. Andrology. 2020 Nov;8(6):1614-1627. doi: 10.1111/andr.12876. Epub 2020 Aug 11. PMID:32737921 doi:http://dx.doi.org/10.1111/andr.12876
- ↑ 16.0 16.1 16.2 16.3 16.4 Kaur H, Werstuck GH. The Effect of Testosterone on Cardiovascular Disease and Cardiovascular Risk Factors in Men: A Review of Clinical and Preclinical Data. CJC Open. 2021 May 17;3(10):1238-1248. doi: 10.1016/j.cjco.2021.05.007. , eCollection 2021 Oct. PMID:34888506 doi:http://dx.doi.org/10.1016/j.cjco.2021.05.007
- ↑ 17.0 17.1 17.2 17.3 17.4 17.5 17.6 Bohl CE, Gao W, Miller DD, Bell CE, Dalton JT. Structural basis for antagonism and resistance of bicalutamide in prostate cancer. Proc Natl Acad Sci U S A. 2005 Apr 26;102(17):6201-6. Epub 2005 Apr 15. PMID:15833816
- ↑ 18.0 18.1 18.2 18.3 18.4 Gao W, Kim J, Dalton JT. Pharmacokinetics and pharmacodynamics of nonsteroidal androgen receptor ligands. Pharm Res. 2006 Aug;23(8):1641-58. doi: 10.1007/s11095-006-9024-3. PMID:16841196 doi:http://dx.doi.org/10.1007/s11095-006-9024-3
- ↑ Sessa F, Salerno M, Di Mizio G, Bertozzi G, Messina G, Tomaiuolo B, Pisanelli D, Maglietta F, Ricci P, Pomara C. Anabolic Androgenic Steroids: Searching New Molecular Biomarkers. Front Pharmacol. 2018 Nov 20;9:1321. doi: 10.3389/fphar.2018.01321. eCollection , 2018. PMID:30524281 doi:http://dx.doi.org/10.3389/fphar.2018.01321
- ↑ 20.0 20.1 20.2 20.3 20.4 20.5 20.6 20.7 Machek SB, Cardaci TD, Wilburn DT, Willoughby DS. Considerations, possible contraindications, and potential mechanisms for deleterious effect in recreational and athletic use of selective androgen receptor modulators (SARMs) in lieu of anabolic androgenic steroids: A narrative review. Steroids. 2020 Dec;164:108753. doi: 10.1016/j.steroids.2020.108753. Epub 2020 Oct , 24. PMID:33148520 doi:http://dx.doi.org/10.1016/j.steroids.2020.108753
- ↑ 21.0 21.1 Fonseca GWPD, Dworatzek E, Ebner N, Von Haehling S. Selective androgen receptor modulators (SARMs) as pharmacological treatment for muscle wasting in ongoing clinical trials. Expert Opin Investig Drugs. 2020 Aug;29(8):881-891. doi: , 10.1080/13543784.2020.1777275. Epub 2020 Jun 18. PMID:32476495 doi:http://dx.doi.org/10.1080/13543784.2020.1777275
- ↑ 22.0 22.1 22.2 Masiello D, Cheng S, Bubley GJ, Lu ML, Balk SP. Bicalutamide functions as an androgen receptor antagonist by assembly of a transcriptionally inactive receptor. J Biol Chem. 2002 Jul 19;277(29):26321-6. doi: 10.1074/jbc.M203310200. Epub 2002 , May 15. PMID:12015321 doi:http://dx.doi.org/10.1074/jbc.M203310200
- ↑ 23.0 23.1 Duke CB, Jones A, Bohl CE, Dalton JT, Miller DD. Unexpected Binding Orientation of Bulky-B-Ring Anti-Androgens and Implications for Future Drug Targets. J Med Chem. 2011 Apr 20. PMID:21506597 doi:10.1021/jm2000097
- ↑ 24.0 24.1 24.2 24.3 Fujita K, Nonomura N. Role of Androgen Receptor in Prostate Cancer: A Review. World J Mens Health. 2019 Sep;37(3):288-295. doi: 10.5534/wjmh.180040. Epub 2018 , Sep 10. PMID:30209899 doi:http://dx.doi.org/10.5534/wjmh.180040
- ↑ 25.0 25.1 Leone G, Tucci M, Buttigliero C, Zichi C, Pignataro D, Bironzo P, Vignani F, Scagliotti GV, Di Maio M. Antiandrogen withdrawal syndrome (AAWS) in the treatment of patients with prostate cancer. Endocr Relat Cancer. 2018 Jan;25(1):R1-R9. doi: 10.1530/ERC-17-0355. Epub 2017 , Sep 28. PMID:28971898 doi:http://dx.doi.org/10.1530/ERC-17-0355
- ↑ 26.0 26.1 26.2 26.3 26.4 26.5 26.6 26.7 26.8 26.9 Cavaliere F, Cozzini P. An insight about the mechanism of action (MoA) of R-bicalutamide on the androgen receptor homodimer using molecular dynamic. Toxicol Appl Pharmacol. 2022 Apr 1;440:115953. doi: 10.1016/j.taap.2022.115953. , Epub 2022 Mar 1. PMID:35245614 doi:http://dx.doi.org/10.1016/j.taap.2022.115953
- ↑ Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik BA, Graff JN, Olmos D, Mainwaring PN, Lee JY, Uemura H, Lopez-Gitlitz A, Trudel GC, Espina BM, Shu Y, Park YC, Rackoff WR, Yu MK, Small EJ. Apalutamide Treatment and Metastasis-free Survival in Prostate Cancer. N Engl J Med. 2018 Apr 12;378(15):1408-1418. doi: 10.1056/NEJMoa1715546. Epub , 2018 Feb 8. PMID:29420164 doi:http://dx.doi.org/10.1056/NEJMoa1715546
- ↑ Pollock Y, Smith MR, Saad F, Chowdhury S, Oudard S, Hadaschik B, Olmos D, Lee JY, Uemura H, Bhaumik A, Londhe A, Rooney B, Brookman-May SD, De Porre P, Mundle SD, Small EJ. Clinical characteristics associated with falls in patients with non-metastatic castration-resistant prostate cancer treated with apalutamide. Prostate Cancer Prostatic Dis. 2022 Oct 8. doi: 10.1038/s41391-022-00592-9. PMID:36209239 doi:http://dx.doi.org/10.1038/s41391-022-00592-9
Proteopedia Page Contributors and Editors (what is this?)
Cristina Benito, Michal Harel, Cristina Murga, Marta Roldan Lazaro, Alexander Berchansky, Joel L. Sussman, Wayne Decatur, David Sánchez Fernández