We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1757
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
== Biological relevance and broader implications == | == Biological relevance and broader implications == | ||
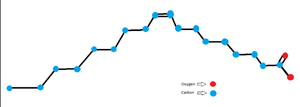
| - | This article’s goal was to test the function and relation of the protein Mevalonate 3,5-bisphosphate and compare it to its homologs. The article seeks to explore the evolutionary changes that the protein has gone through that managed to separate it from its suspected evolutionary predecessors which require ATP to function and allowed it to become ATP-independent. This suspected evolutionary relevance is due to the residual signs of the ATP binding site which are still present in the Mevalonate 3,5-bisphosphate despite the protein being completely ATP-independent. | + | This article’s goal was to test the function and relation of the protein Mevalonate 3,5-bisphosphate and compare it to its homologs. The article seeks to explore the evolutionary changes that the protein has gone through that managed to separate it from its suspected evolutionary predecessors which require ATP to function and allowed it to become ATP-independent. This suspected evolutionary relevance is due to the residual signs of the ATP binding site which are still present in the Mevalonate 3,5-bisphosphate despite the protein being completely ATP-independent. This ATP binding site present in Mevalonate 3,5-bisphosphate's likely evolutionary predecessors has been bound to by a ligand similar to a fatty acid. |
[[Image:Oleic Acid.png|300 px|thumb|right|Oleic Acid(suspected ligand present)]] | [[Image:Oleic Acid.png|300 px|thumb|right|Oleic Acid(suspected ligand present)]] | ||
== Important amino acids== | == Important amino acids== | ||
| Line 14: | Line 14: | ||
Mevalonate 3,5-bisphosphate is a homodimer made up of <scene name='93/934001/Secondary/1'>40% alpha helices and 60% beta-sheets</scene>. Because Mevalonate 3,5-bisphosphate is a homodimer both molecules present are <scene name='93/934001/Quat/1'>identical</scene> in primary, secondary, and tertiary structure. Mevalonate 3,5-bisphosphate is amphipathic as showcased by the surplus of hydrophobic as well as hydrophilic amino acids present in its <scene name='93/934001/Active_site/2'>Active Site</scene>. This<scene name='93/934001/Active_site/4'> view</scene> also shows us the large cleft for substrate binding present on the molecule. | Mevalonate 3,5-bisphosphate is a homodimer made up of <scene name='93/934001/Secondary/1'>40% alpha helices and 60% beta-sheets</scene>. Because Mevalonate 3,5-bisphosphate is a homodimer both molecules present are <scene name='93/934001/Quat/1'>identical</scene> in primary, secondary, and tertiary structure. Mevalonate 3,5-bisphosphate is amphipathic as showcased by the surplus of hydrophobic as well as hydrophilic amino acids present in its <scene name='93/934001/Active_site/2'>Active Site</scene>. This<scene name='93/934001/Active_site/4'> view</scene> also shows us the large cleft for substrate binding present on the molecule. | ||
== Other important features == | == Other important features == | ||
| - | + | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<ref>Aoki, M., Vinokur, J., Motoyama, K., Ishikawa, R., Collazo, M., Cascio, D., Sawaya, M. R., Ito, T., Bowie, J. U., & Hemmi, H. (2022). Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways. The Journal of Biological Chemistry, 298(7), 102111–102111. https://doi.org/10.1016/j.jbc.2022.102111</ref> | <ref>Aoki, M., Vinokur, J., Motoyama, K., Ishikawa, R., Collazo, M., Cascio, D., Sawaya, M. R., Ito, T., Bowie, J. U., & Hemmi, H. (2022). Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways. The Journal of Biological Chemistry, 298(7), 102111–102111. https://doi.org/10.1016/j.jbc.2022.102111</ref> | ||
| - | <ref>Vinokur, J M, et al. “7T71: Crystal Structure of Mevalonate 3,5-Bisphosphate Decarboxylase from Picrophilus Torridus.” RCSB PDB, https://www.rcsb.org/structure/7t71.</ref> | ||
<references/> | <references/> | ||
Revision as of 16:59, 13 December 2022
| This Sandbox is Reserved from November 4, 2022 through January 1, 2023 for use in the course CHEM 351 Biochemistry taught by Bonnie Hall at the Grand View University, Des Moines, USA. This reservation includes Sandbox Reserved 1755 through Sandbox Reserved 1764. |
To get started:
More help: Help:Editing |
Mevalonate 3,5-bisphosphate decarboxylase
| |||||||||||
References
[1]
- ↑ Aoki, M., Vinokur, J., Motoyama, K., Ishikawa, R., Collazo, M., Cascio, D., Sawaya, M. R., Ito, T., Bowie, J. U., & Hemmi, H. (2022). Crystal structure of mevalonate 3,5-bisphosphate decarboxylase reveals insight into the evolution of decarboxylases in the mevalonate metabolic pathways. The Journal of Biological Chemistry, 298(7), 102111–102111. https://doi.org/10.1016/j.jbc.2022.102111