User:Katherine Snook/Sandbox 1
From Proteopedia
< User:Katherine Snook(Difference between revisions)
| Line 7: | Line 7: | ||
[https://www.rcsb.org/structure/7UPI Holoenzyme Structure] | [https://www.rcsb.org/structure/7UPI Holoenzyme Structure] | ||
<scene name='95/954076/Random_selection_10-100/5'>Residues 10-100 white</scene> | <scene name='95/954076/Random_selection_10-100/5'>Residues 10-100 white</scene> | ||
| + | <ref name= "Kwon">PMID:35831509</ref> | ||
===active site=== | ===active site=== | ||
| - | + | <ref name="Ransey">PMID:28504306</ref> | |
===binding pocket=== | ===binding pocket=== | ||
Current revision
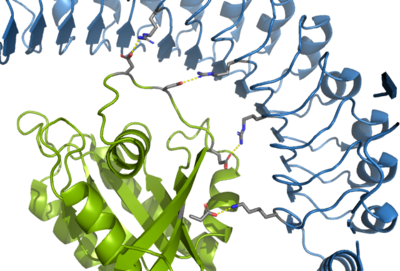
SHOC2-MRAS-PP1C holoenzyme
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Kwon JJ, Hajian B, Bian Y, Young LC, Amor AJ, Fuller JR, Fraley CV, Sykes AM, So J, Pan J, Baker L, Lee SJ, Wheeler DB, Mayhew DL, Persky NS, Yang X, Root DE, Barsotti AM, Stamford AW, Perry CK, Burgin A, McCormick F, Lemke CT, Hahn WC, Aguirre AJ. Structure-function analysis of the SHOC2-MRAS-PP1C holophosphatase complex. Nature. 2022 Jul 13. pii: 10.1038/s41586-022-04928-2. doi:, 10.1038/s41586-022-04928-2. PMID:35831509 doi:http://dx.doi.org/10.1038/s41586-022-04928-2
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
- Emily Landwehr
- Reagann Meyers