We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1768
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
=== Significant Residues === | === Significant Residues === | ||
| - | The majority of residues involved in bile salt uptake are also involved in HBV/HDV infection. <scene name='95/952696/Residues_84-87_1/1'>Residues 84-87</scene> (extracellular view) of Human NTCP have been shown to be vital for HBV/HDV virus recognition along with bile salt uptake. These residues were replaced in mice NTCP by human NTCP and conferred to successful binding of the virus. These residues are found in the extracellular loop connecting TM2 and TM3.<Ref name="Park"> Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. [https://dx.doi.org/10.1038/s41586-022-04857-0 DOI: 10.1038/s41586-022-04857-0]. </Ref> <scene name='95/952696/Residues_157-165/1'>Residues 157-165</scene> (extracellular view) have also been shown to be vital for HBV/HDV viral recognition and bile salt uptake. These residues were mutated in monkey NTCP to the human residues and preS1 binding was then successful. These residues are found on the N-terminal end of TM5. The absence of residues in either of these <scene name='95/952696/Residues_84-87_and_157-165/1'>two extracellular patches</scene> hinders preS1 binding and therefore HBV/HDV infection. Interestingly, residues 84-87 do not affect bile acid uptake, so it is a potential site for blocking HBV/HDV infection while maintaining NTCP's ability to perform its normal function.<ref name="Park" /> Another important residue was discovered to be a [https://en.wikipedia.org/wiki/Single-nucleotide_polymorphism single-nucleotide polymorphism] in a small population in East Asia. <scene name='95/952696/Residue_267/1'>Residue 267</scene>, which is normally serine, being mutated to phenylalanine prevents preS1 binding and does not support bile acid transport. This residue is also found extracellularly, on TM8 of NTCP.<ref name="Qi" /> There are 3 additional leucine residues that when mutated, block both preS1 binding and HBV/HDV infection. Replacing L27, L31, and L35 | + | The majority of residues involved in bile salt uptake are also involved in HBV/HDV infection. <scene name='95/952696/Residues_84-87_1/1'>Residues 84-87</scene> (extracellular view) of Human NTCP have been shown to be vital for HBV/HDV virus recognition along with bile salt uptake. These residues were replaced in mice NTCP by human NTCP and conferred to successful binding of the virus. These residues are found in the extracellular loop connecting TM2 and TM3.<Ref name="Park"> Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. [https://dx.doi.org/10.1038/s41586-022-04857-0 DOI: 10.1038/s41586-022-04857-0]. </Ref> <scene name='95/952696/Residues_157-165/1'>Residues 157-165</scene> (extracellular view) have also been shown to be vital for HBV/HDV viral recognition and bile salt uptake. These residues were mutated in monkey NTCP to the human residues and preS1 binding was then successful. These residues are found on the N-terminal end of TM5. The absence of residues in either of these <scene name='95/952696/Residues_84-87_and_157-165/1'>two extracellular patches</scene> hinders preS1 binding and therefore HBV/HDV infection. Interestingly, residues 84-87 do not affect bile acid uptake, so it is a potential site for blocking HBV/HDV infection while maintaining NTCP's ability to perform its normal function.<ref name="Park" /> Another important residue was discovered to be a [https://en.wikipedia.org/wiki/Single-nucleotide_polymorphism single-nucleotide polymorphism] in a small population in East Asia. <scene name='95/952696/Residue_267/1'>Residue 267</scene>, which is normally serine, being mutated to phenylalanine prevents preS1 binding and does not support bile acid transport. This residue is also found extracellularly, on TM8 of NTCP.<ref name="Qi" /> There are 3 additional leucine residues that when mutated, block both preS1 binding and HBV/HDV infection. Replacing <scene name='95/952696/Leucine_residues/1'>L27, L31, and L35</scene> with tryptophan residues presumably blocks the preS1 binding site preventing proper infection.<ref name="Park" /> |
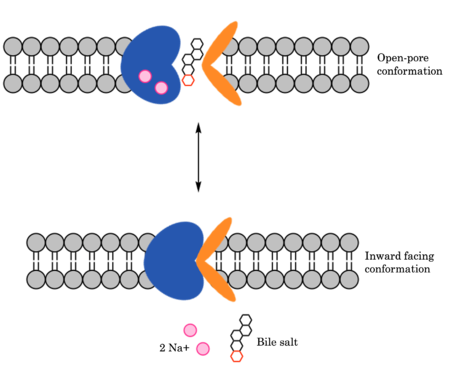
== Function == | == Function == | ||
Revision as of 21:51, 29 March 2023
Sodium-taurocholate Co-transporting Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. DOI: 10.1038/s41586-022-04723-z.
- ↑ 2.0 2.1 Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. DOI: 10.1038/s41586-022-04845-4.
- ↑ Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014 Jul;35(23):6130-41. doi: 10.1016/j.biomaterials.2014.04.037. DOI: 10.1016/j.biomaterials.2014.04.037.
- ↑ 4.0 4.1 Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. doi: 10.1016/j.xinn.2022.100294. DOI: 10.1016/j.xinn.2022.100294.
- ↑ 5.0 5.1 Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. DOI: 10.1038/s41422-022-00680-4.
- ↑ 6.0 6.1 6.2 Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. DOI: 10.1038/s41586-022-04857-0.
Student Contributors
- Ben Minor
- Maggie Samm
- Zac Stanley