We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1769
From Proteopedia
(Difference between revisions)
| Line 4: | Line 4: | ||
== Introduction == | == Introduction == | ||
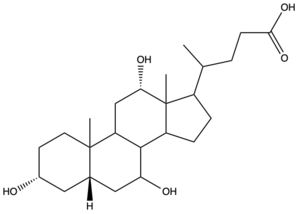
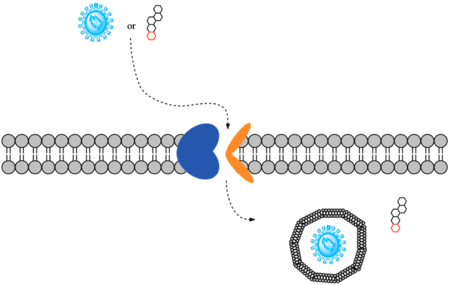
Sodium-taurocholate Co-transporting Polypeptide (NTCP) is a member of the solute carrier membrane transport proteins. It is found within the membrane of [[Image:Bile Salt.png|300 px|right|thumb|Figure 1. Structure of Cholic Acid, an example of a bile acid.]][https://en.wikipedia.org/wiki/Hepatocyte hepatocytes], and its primary role is to facilitate the transport of [https://en.wikipedia.org/wiki/Bile_acid bile salts] (Figure 1) into hepatocytes from the bloodstream.<Ref name="Goutam"> Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. [https://dx.doi.org/10.1038/s41586-022-04723-z DOI: 10.1038/s41586-022-04723-z]. </Ref> This is important because 90% of human bile salts are recycled daily, so the function of NTCP is critical in providing bile acids to solubilize fats for digestion. In addition to transporting bile acids into the cytoplasm of hepatocytes, NTCP also serves as a receptor for [https://en.wikipedia.org/wiki/Hepatitis_B Hepatitis B (HBV)] and [https://en.wikipedia.org/wiki/Hepatitis_D Hepatitis D (HDV)] viruses.<Ref name="Asami"> Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. [https://dx.doi.org/10.1038/s41586-022-04845-4 DOI: 10.1038/s41586-022-04845-4]. </Ref> [[Image:Basic_mech_NTCP.png|450 px|thumb|Figure 2. Overall mechanism of NTCP.]] | Sodium-taurocholate Co-transporting Polypeptide (NTCP) is a member of the solute carrier membrane transport proteins. It is found within the membrane of [[Image:Bile Salt.png|300 px|right|thumb|Figure 1. Structure of Cholic Acid, an example of a bile acid.]][https://en.wikipedia.org/wiki/Hepatocyte hepatocytes], and its primary role is to facilitate the transport of [https://en.wikipedia.org/wiki/Bile_acid bile salts] (Figure 1) into hepatocytes from the bloodstream.<Ref name="Goutam"> Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. [https://dx.doi.org/10.1038/s41586-022-04723-z DOI: 10.1038/s41586-022-04723-z]. </Ref> This is important because 90% of human bile salts are recycled daily, so the function of NTCP is critical in providing bile acids to solubilize fats for digestion. In addition to transporting bile acids into the cytoplasm of hepatocytes, NTCP also serves as a receptor for [https://en.wikipedia.org/wiki/Hepatitis_B Hepatitis B (HBV)] and [https://en.wikipedia.org/wiki/Hepatitis_D Hepatitis D (HDV)] viruses.<Ref name="Asami"> Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. [https://dx.doi.org/10.1038/s41586-022-04845-4 DOI: 10.1038/s41586-022-04845-4]. </Ref> [[Image:Basic_mech_NTCP.png|450 px|thumb|Figure 2. Overall mechanism of NTCP.]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
== Structure == | == Structure == | ||
| - | + | NTCP is a transmembrane protein found in hepatocyte cells. It consists of nine [https://en.wikipedia.org/wiki/Alpha_helix alpha helices] spanning the membrane, with the N-terminus located on the extracellular side of the plasma membrane and the C-terminus located on the intracellular side (FIGURE 3 - TOPOGRAPHY PIC). Transmembrane helices are connected by short loops as well as extracellular and intracellular alpha helices that lie nearly parallel to the membrane.<ref name="Asami" /> Structures were determined by [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy cryogenic electron microscopy (Cryo-EM)] of NTCP in complex with antibodies or nanobodies. (CITE 4 PAPERS HERE) | |
=== Domains === | === Domains === | ||
| - | NTCP contains two characteristic domains: the core and panel domains. Movement of these two domains allows recognition and transport of bile | + | NTCP contains two characteristic domains (INSERT GREEN LINK): the core and panel domains. Movement of these two domains allows recognition and transport of bile acids into hepatocytes. |
| - | *<b><font color="orange">Panel Domain</font></b>: Residues 1-44, 155-208 | + | *<b><font color="orange">Panel Domain</font></b> (INSERT GREEN LINK making core more transparent): Residues 1-44, 155-208 |
** Formed by transmembrane helices TM1, TM5, and TM6. | ** Formed by transmembrane helices TM1, TM5, and TM6. | ||
| - | *<b><font color="#0040e0">Core domain</font></b>: Residues 45-154, 209-309 | + | *<b><font color="#0040e0">Core domain</font></b> (INSERT GREEN LINK making panel more transparent): Residues 45-154, 209-309 |
**Formed by the packing of a helix bundle of TM2, TM3, and TM4 with another helix bundle of TM7, TM8, and TM9. These two helix bundles are related by pseudo two-fold symmetry.<Ref name="Qi"> Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. [https://dx.doi.org/10.1016/j.xinn.2022.100294 DOI: 10.1016/j.xinn.2022.100294]. </Ref> | **Formed by the packing of a helix bundle of TM2, TM3, and TM4 with another helix bundle of TM7, TM8, and TM9. These two helix bundles are related by pseudo two-fold symmetry.<Ref name="Qi"> Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. [https://dx.doi.org/10.1016/j.xinn.2022.100294 DOI: 10.1016/j.xinn.2022.100294]. </Ref> | ||
=== Proline/Glycine Hinge === | === Proline/Glycine Hinge === | ||
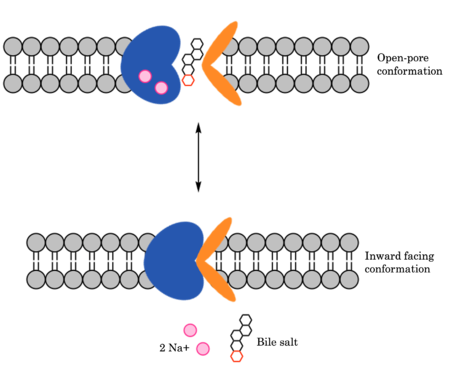
| - | Glycine and proline residues in the connecting loops and extra- and intracellular helices act as hinges in the mechanism of bile salt uptake. | + | Glycine and proline residues (INSERT GREEN LINK) in the connecting loops and extra- and intracellular helices (Figure 3) act as hinges in the mechanism of bile salt uptake. This flexibility allows separation of the core and panel domains, creating a pore open to the extracellular space and exposing critical Na+ binding sites. Once substrate binds the open-pore state, this hinge allows the transition to close this pore relative to the extracellular side and open to the cytoplasmic side, thus allowing release of substrate into the cell.<ref name = "Goutam" /> |
=== Sodium Binding Sites === | === Sodium Binding Sites === | ||
| - | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions are required to be bound to to NTCP in the open-pore state.<Ref name="Liu"> Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. [https://dx.doi.org/10.1038/s41422-022-00680-4 DOI: 10.1038/s41422-022-00680-4]. </Ref> | + | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions are required to be bound to to NTCP in the open-pore state.<Ref name="Liu"> Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. [https://dx.doi.org/10.1038/s41422-022-00680-4 DOI: 10.1038/s41422-022-00680-4]. </Ref> The residues in the first sodium binding include <scene name='95/952697/Sodium_binding_sites1/5'>Ser105, Asn106, Thr 123, and Glu 257</scene>. The residues in the second sodium binding site include <scene name='95/952697/Sodium_binding_sites2/3'>Gln68 and Gln261</scene>. Mutations to these significant residues inhibit the binding of sodium ions, and consequently, inhibit the transport of bile salts by NTCP.<ref name = "Liu" />[https://en.wikipedia.org/wiki/Active_transport#Secondary_active_transport secondary active transport] is used here, as the transport of bile acids into the cell is so thermodynamically unfavorable that the reaction has to be coupled to the favorable transport of two sodium into into the cell.<ref name = "Goutam" /> . When the bile salts are released into the cell, the protein is then found in the inward facing conformation, in which the pore through which the it had just passed is now closed to the extracellular side. |
=== Significant Residues === | === Significant Residues === | ||
Revision as of 15:54, 30 March 2023
Sodium-taurocholate Co-transporting Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. DOI: 10.1038/s41586-022-04723-z.
- ↑ 2.0 2.1 Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. DOI: 10.1038/s41586-022-04845-4.
- ↑ 3.0 3.1 Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. DOI: 10.1016/j.xinn.2022.100294.
- ↑ 4.0 4.1 Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. DOI: 10.1038/s41422-022-00680-4.
- ↑ 5.0 5.1 5.2 5.3 Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. DOI: 10.1038/s41586-022-04857-0.
- ↑ 6.0 6.1 Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014 Jul;35(23):6130-41. DOI: 10.1016/j.biomaterials.2014.04.037.
- ↑ Patton JS, Carey MC. Watching fat digestion. Science. 1979 Apr 13;204(4389):145-8. DOI: 10.1126/science.432636.
Student Contributors
- Ben Minor
- Maggie Samm
- Zac Stanley