Background
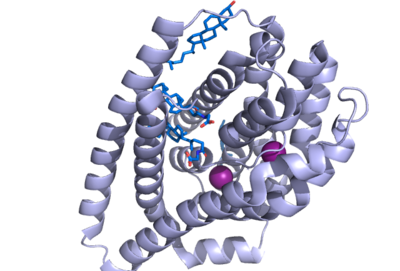
Figure 1. NTCP structure with both Na ions and bile salts bound. PDB file 7ZYI.
Sodium taurocholate co-transporting polypeptide (NTCP) is a sodium-dependent transporter in the body that is responsible for the transportation of bile salts from the blood into epithelial liver cells. This carrier protein is responsible for a conformational change that allows bile salts to cross the cell membrane and enter the inside of liver cells. [1]. Both sodium ions and bile salts bind to NTCP in the same binding pocket on the molecule (Fig. 1).
NTCP also acts as a receptor for Hepatitis B virus and Hepatitis D virus. The bile salts transported by NTCP are located within the gastrointestinal tract of the body and play a very key role in many biological functions. These functions include digesting and absorbing nutrients by helping break down fats and transporting lipid soluble nutrients into the liver. [2]. The NTCP carrier protein itself can be found within hepatocytes, or the epithelial cells of the liver, but more specifically, within the basolateral membrane of these cells. [3]. The uptake of bile salts into the liver also allow for drugs to be both absorbed and excreted, as well as essential nutrients such as Vitamin A,D,E, and K to be absorbed in the small intestine.
Structural Overview
NTCP has 9 transmembrane alpha helices (TM) that form the protein, with an extracellular N-terminus. NTCP has two domains within the protein, a panel domain, made up of TM1, TM5, and TM6, and a core domain, made up of TM2-4 and TM7-9. An interesting feature of NTCP is the cross of TM3 and TM8 that form an X within the protein. The two domains are essential to the conformation change of NTCP to bind bile salts. [1]
There are two significant areas in the NTCP structure that facilitate ligand binding, which are referred to as "patches." Residues 84-87 of NTCP are patch 1, which are located on the TM2-TM3 loop in the core domain. This is also considered the extracellular region of NTCP “tunnel." Residues 157-165 NTCP are associated with patch 2. They are located on N-terminal half of the TM5 in the panel domain (residue sequence: KGIVISLVL). Patch 2 is also located in th extracellular region. These residues' importance was determined through mutations of these residues and examined through pull-down assays (Asami, et. al, 2022).
Binding Pocket
The NTCP binding pocket represents a "tunnel" lined with hydrophilic residues within the NTCP structure to allow hydrophilic bile salts and sodium ions to bind. When there are no bile salts bound to NTCP's binding tunnel, it forms a hollow hole in the middle of the structure, from to . As bile salts bind to this binding pocket, the bile salts very nicely fill the hole within NTCP and fit almost perfectly within the tunnel, to the point where the can no longer be seen, as it is completely encompassing the bile salts. This tunnel formed connects the external cytoplasm of the hepatocyte to the inner leaflet of the basolateral membrane. The side of the tunnel where the bile salts bind are lined with hydrophilic residues, whereas the opposite side of the helix is lined with hydrophobic residues, as the transmembrane helices are amphipathic. [4].
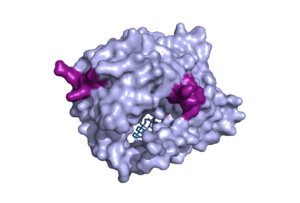
Figure 1. Surface representation of the NTCP molecule with both patches shown. Patch 1 can be seen on the left side of the molecule, whereas patch 2 is located on the right side within the binding tunnel. PDB file 7ZYI.
The binding pocket of NTCP contains two patches on its exterior. These two patches are used for assisting in the binding of bile salts into the binding tunnel. These external patches are extremely important for aiding in the transport of bile salts from the exterior of NTCP to the interior binding tunnel [3]. Patch 1 is made up of residues 84-87 of NTCP and Patch 2 consists of residues 157-165. Patch 1 is located on the poles of NTCP, namely the top of the structure, within the TM2-TM3 transmembrane loop, whereas patch 2 is located towards the middle of the NTCP molecule within transmembrane 5 (TM5). These two patches are also predominantly responsible for binding the preS1 binding region of the HBV/HDV virus. Patch 2 also forms a part of the binding tunnel previously mentioned [3].
Conformation Change
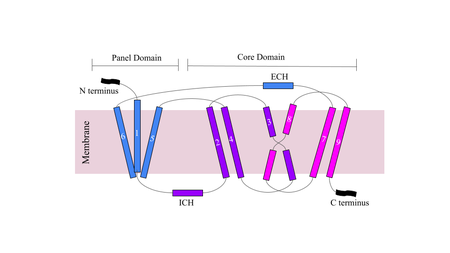
Figure 1. Cartoon of NTCP topology.
The conformational change of NTCP's core domain helices are essential to bile salt binding and uptake. Figure 1 displays the topology of NTCP, highlighting both the panel (shown in blue) and core (shown in purple and pink) domains. Helices 3 and 8 are the main structural components of the conformational change. Before bile salt can bind, the pore in which salt binds must be . The pore is flipped toward the outer membrane to allow for binding. Once , the pore is , and bile salt is able to be released into the cell, past the inner membrane.
Mechanism

Figure 1. Bile Salt Uptake Mechanism.
The NTCP protein goes through a conformational change when assisting in the uptake of bile salt into the cell. This is accomplished through the opening of a wide transmembrane pore, creating a transport pathway for bile salts. The mechanism includes two metal ions that allow for residue stabilization when going through the conformational change. Binding of the preS1 region of the HBV/HDV virus blocks any subsequent bile salt uptake. Thus, preS1 binding blocks the conformational change and entry of any salts into the cell. Residues 8-17 of preS1 are critical for NTCP:pres1 binding. Patch 1 and Patch 2 (external) residues interact with residues 8-17 of preS1 to facilitate binding.
Significance
NTCP serves a multitude of biological functions, including bile salt uptake and HBV/HDV binding. NTCP is a key player in the absorption and digestion of fats and fat-soluble vitamins in the body, as well as the synthesis of bile within the liver. The uptake of bile salts, transcriptional and post-transcriptional, are signaling molecules for the liver and intestine.
Bile Salt Uptake
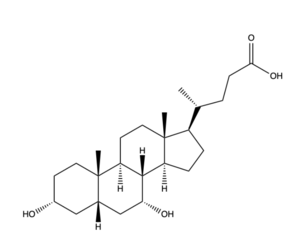
Figure 2. Bile Salt Structure.
Disease of the liver is due to a decrease in bile salt uptake. This disease is transferred through bodily fluids between organisms. Liver Disease causes symptoms such as jaundice, abdominal pain and swelling, and swelling of the legs and feet. Liver disease can lead to liver cancer and other life-threatening diseases, making bile salt uptake essential to liver function.
HBV/HDV
Hepatitis B and D both rely on NTCP to bind and infect a human. HBV/HDV uses a similar mechanism as bile salt uptake within NTCP. The myristoylated (myr) pre-S1 domain of HBV is critical for its binding within the protein. Residues 8-17 within the pre-S1 domain were found to be most important in pre-S1 binding to NTCP. HBV/HDV uses the binding patches on the outside of the protein to successfully bind. HBV/HDV binding halts the uptake of bile salt, indicating that the tunnel formed within NTCP allows the uptake of bile salts and may also mediate the binding of HBV/HDV. [[2]]





