We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1769
From Proteopedia
(Difference between revisions)
| Line 21: | Line 21: | ||
=== Sodium Binding Sites === | === Sodium Binding Sites === | ||
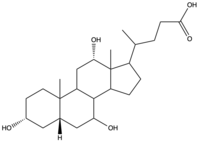
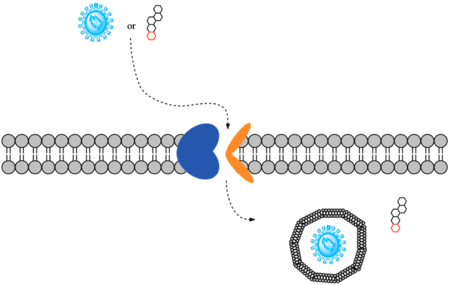
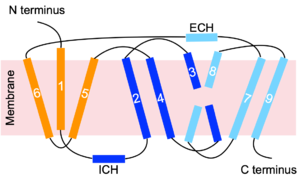
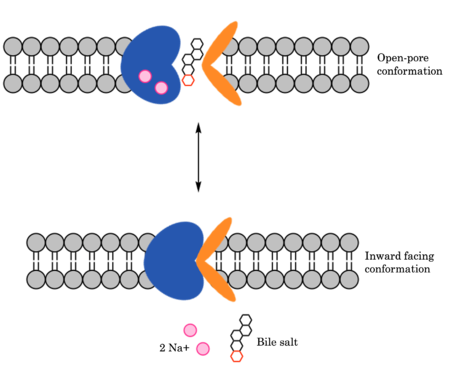
| - | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions are required to be bound to to NTCP in the open-pore state.<Ref name="Liu"> Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. [https://dx.doi.org/10.1038/s41422-022-00680-4 DOI: 10.1038/s41422-022-00680-4]. </Ref> Thus, there are two <scene name='95/952697/Ntcp_complex_sodiumsites/4'>sodium binding sites</scene>. The residues in the <scene name='95/952697/Sodium_binding_sites1/ | + | To transport a single bile salt from the blood to the cytoplasm of the hepatocyte, two sodium ions are required to be bound to to NTCP in the open-pore state.<Ref name="Liu"> Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. [https://dx.doi.org/10.1038/s41422-022-00680-4 DOI: 10.1038/s41422-022-00680-4]. </Ref> Thus, there are two <scene name='95/952697/Ntcp_complex_sodiumsites/4'>sodium binding sites</scene>. The residues in the <scene name='95/952697/Sodium_binding_sites1/7'>first sodium binding site</scene> include Ser105, Asn106, Thr123, and Glu257. The residues in the <scene name='95/952697/Sodium_binding_sites2/3'>second sodium binding site</scene> include Gln68 and Gln261. Mutations to these significant residues inhibit the binding of sodium ions, and consequently, inhibit the transport of bile salts by NTCP.<ref name = "Liu" /> [https://en.wikipedia.org/wiki/Active_transport#Secondary_active_transport Secondary active transport] is used here, as the transport of bile acids into the cell is so thermodynamically unfavorable that the reaction has to be coupled to the favorable transport of two sodium into into the cell.<ref name = "Goutam" /> . When the bile salts are released into the cell, the protein is then found in the inward facing conformation, in which the pore through which the it had just passed is now closed to the extracellular side. |
=== Significant Residues for HBV/HDV Infection === | === Significant Residues for HBV/HDV Infection === | ||
Revision as of 21:16, 2 April 2023
Sodium-taurocholate Co-transporting Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Goutam K, Ielasi FS, Pardon E, Steyaert J, Reyes N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature. 2022 Jun;606(7916):1015-1020. DOI: 10.1038/s41586-022-04723-z.
- ↑ 2.0 2.1 2.2 2.3 Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. DOI: 10.1038/s41586-022-04845-4.
- ↑ 3.0 3.1 3.2 3.3 3.4 Park JH, Iwamoto M, Yun JH, Uchikubo-Kamo T, Son D, Jin Z, Yoshida H, Ohki M, Ishimoto N, Mizutani K, Oshima M, Muramatsu M, Wakita T, Shirouzu M, Liu K, Uemura T, Nomura N, Iwata S, Watashi K, Tame JRH, Nishizawa T, Lee W, Park SY. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature. 2022 Jun;606(7916):1027-1031. DOI: 10.1038/s41586-022-04857-0.
- ↑ 4.0 4.1 4.2 Liu H, Irobalieva RN, Bang-Sørensen R, Nosol K, Mukherjee S, Agrawal P, Stieger B, Kossiakoff AA, Locher KP. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 2022 Aug;32(8):773-776. DOI: 10.1038/s41422-022-00680-4.
- ↑ 5.0 5.1 Qi X, Li W. Unlocking the secrets to human NTCP structure. Innovation (Camb). 2022 Aug 1;3(5):100294. DOI: 10.1016/j.xinn.2022.100294.
- ↑ 6.0 6.1 Zhang X, Zhang Q, Peng Q, Zhou J, Liao L, Sun X, Zhang L, Gong T. Hepatitis B virus preS1-derived lipopeptide functionalized liposomes for targeting of hepatic cells. Biomaterials. 2014 Jul;35(23):6130-41. DOI: 10.1016/j.biomaterials.2014.04.037.
- ↑ Patton JS, Carey MC. Watching fat digestion. Science. 1979 Apr 13;204(4389):145-8. DOI: 10.1126/science.432636.
- ↑ Donkers JM, Kooijman S, Slijepcevic D, Kunst RF, Roscam Abbing RL, Haazen L, de Waart DR, Levels JH, Schoonjans K, Rensen PC, Oude Elferink RP, van de Graaf SF. NTCP deficiency in mice protects against obesity and hepatosteatosis. JCI Insight. 2019 Jun 25;5(14):e127197. DOI: 10.1172/jci.insight.127197.
Student Contributors
- Ben Minor
- Maggie Samm
- Zac Stanley