Phosphofructokinase (PFK)
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
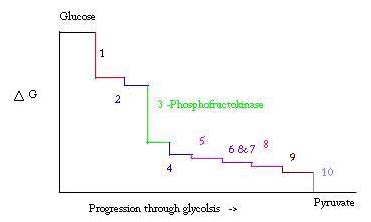
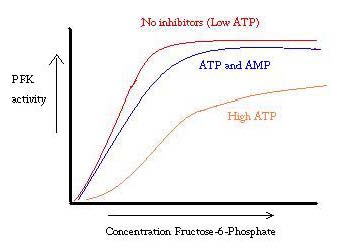
'''Phosphofructokinase-1''' (PFK-1) is a glycolytic enzyme that catalyzes the transfer of a phosphoryl group from <scene name='Phosphofructokinase_(PFK)/Cv/21'>ATP</scene> to <scene name='Phosphofructokinase_(PFK)/Cv/22'>fructose-6-phosphate (F6P)</scene> to yield <scene name='Phosphofructokinase_(PFK)/Cv/16'>ADP</scene> and <scene name='Phosphofructokinase_(PFK)/Cv/23'>fructose-1,6-bisphosphate (FBP)</scene>. See [[Glycolysis Enzymes]]. Mg2+ is also important in this reaction (<scene name='Phosphofructokinase_(PFK)/Cv/24'>click here to see animation of reaction</scene>). '''Phosphofructokinase-2''' (PFK-2) acts on the same substrates to yield ADP and <scene name='Phosphofructokinase_(PFK)/Cv1/3'>fructose-2,6-bisphosphate (F2,6P)</scene>. <scene name='Phosphofructokinase_(PFK)/Cv1/4'>Click here to see the difference between FBP and F2,6P</scene>. PFK reaction is strongly exergonic (irreversible) under physiological conditions and hence is one of the glycolytic pathway's rate-determining steps. In most organisms/tissues, PFK is the glycolytic pathway's major flux-regulating enzyme; its activity is controlled by the concentrations of an unusually large number of metabolites including ATP, ADP, AMP, PEP and fructose-2,6-bisphosphate. | '''Phosphofructokinase-1''' (PFK-1) is a glycolytic enzyme that catalyzes the transfer of a phosphoryl group from <scene name='Phosphofructokinase_(PFK)/Cv/21'>ATP</scene> to <scene name='Phosphofructokinase_(PFK)/Cv/22'>fructose-6-phosphate (F6P)</scene> to yield <scene name='Phosphofructokinase_(PFK)/Cv/16'>ADP</scene> and <scene name='Phosphofructokinase_(PFK)/Cv/23'>fructose-1,6-bisphosphate (FBP)</scene>. See [[Glycolysis Enzymes]]. Mg2+ is also important in this reaction (<scene name='Phosphofructokinase_(PFK)/Cv/24'>click here to see animation of reaction</scene>). '''Phosphofructokinase-2''' (PFK-2) acts on the same substrates to yield ADP and <scene name='Phosphofructokinase_(PFK)/Cv1/3'>fructose-2,6-bisphosphate (F2,6P)</scene>. <scene name='Phosphofructokinase_(PFK)/Cv1/4'>Click here to see the difference between FBP and F2,6P</scene>. PFK reaction is strongly exergonic (irreversible) under physiological conditions and hence is one of the glycolytic pathway's rate-determining steps. In most organisms/tissues, PFK is the glycolytic pathway's major flux-regulating enzyme; its activity is controlled by the concentrations of an unusually large number of metabolites including ATP, ADP, AMP, PEP and fructose-2,6-bisphosphate. | ||
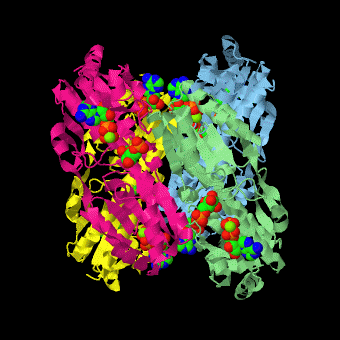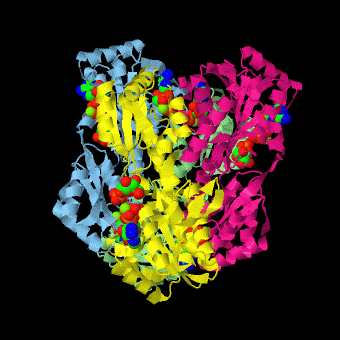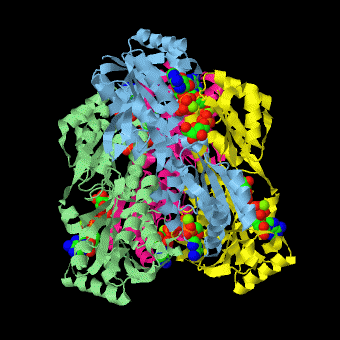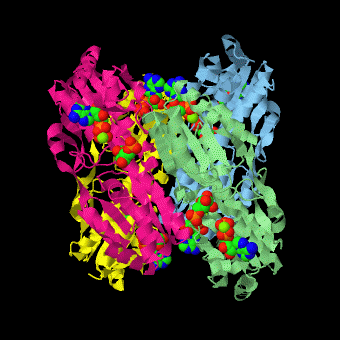
| - | <scene name='Phosphofructokinase_(PFK)/4pfk_biol/3'>PFK from B. stearothermophilus</scene> is a tetramer of identical 320-residue subunits. It is an allosteric enzyme that is described using the symmetry model of allosterism whereby there is a concerted transition from its high-activity R state to its low-activity T state. The X-ray structures of both R and T states of the enzyme have been reported.<ref>PMID:2136935</ref> The binding of one molecule of its substrate F6P, which binds to the R state enzyme with high affinity but to the T state enzyme with low affinity, causes PFK to take up the R state, which in turn, increases the binding affinity of the enzyme for additional F6P (a homotropic effect). Activators, such as ADP and AMP bind to so-called allosteric sites, binding sites distinct from the active site, where they likewise facilitate the formation of the R state and hence activate the enzyme (a heterotropic effect; ADP, being a product of the PFK reaction, also binds at the enzyme's active site). Similarly, inhibitors such as PEP bind to allosteric sites (which in the case of PFK overlaps the activating allosteric site) where they promote the formation of the T state, thereby inhibiting the enzyme. | + | <scene name='Phosphofructokinase_(PFK)/4pfk_biol/3'>PFK from B. stearothermophilus</scene> is a tetramer of identical 320-residue subunits. It is an allosteric enzyme that is described using the symmetry model of allosterism whereby there is a concerted transition from its high-activity R state to its low-activity T state. The X-ray structures of both R and T states of the enzyme have been reported.<ref>PMID:2136935</ref> The binding of one molecule of its substrate <scene name='37/376373/F6p/1'>F6P</scene>, which binds to the R state enzyme with high affinity but to the T state enzyme with low affinity, causes PFK to take up the R state, which in turn, increases the binding affinity of the enzyme for additional F6P (a homotropic effect). Activators, such as <scene name='37/376373/Allosteric_adp/1'>ADP</scene> and AMP bind to so-called allosteric sites, binding sites distinct from the active site, where they likewise facilitate the formation of the R state and hence activate the enzyme (a heterotropic effect; ADP, being a product of the PFK reaction, also binds at the enzyme's active site). Similarly, inhibitors such as PEP bind to allosteric sites (which in the case of PFK overlaps the activating allosteric site) where they promote the formation of the T state, thereby inhibiting the enzyme. |
Two of the active sites of the enzyme are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ad_overview/1'>subunits A (light blue) and D (yellow)</scene> with the active site interfaces in magenta with the substrates in cyan. Two more active sites are at the interface of subunits B (green) and C (pink). A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ad_closeup/1'>A/D interface active site</scene> of subunit D (Yellow) shows that amino acids from both subunits A (light blue) and D (Yellow) contribute to the binding of F6P. Two of the allosteric sites are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ab_overview/1'>subunits A and B</scene> and two at the interface of subunits C and D. Again the interfaces are magenta with the allosteric ligand in cyan. A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ab_closeup/1'>A/B allosteric site</scene> of subunit A shows contributions from both subunits to the binding of ADP. The conformational changes in going between the R and T states of PFK are illustrated below. | Two of the active sites of the enzyme are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ad_overview/1'>subunits A (light blue) and D (yellow)</scene> with the active site interfaces in magenta with the substrates in cyan. Two more active sites are at the interface of subunits B (green) and C (pink). A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ad_closeup/1'>A/D interface active site</scene> of subunit D (Yellow) shows that amino acids from both subunits A (light blue) and D (Yellow) contribute to the binding of F6P. Two of the allosteric sites are located at the interface of <scene name='Phosphofructokinase_(PFK)/Pfk_ab_overview/1'>subunits A and B</scene> and two at the interface of subunits C and D. Again the interfaces are magenta with the allosteric ligand in cyan. A closeup of the <scene name='Phosphofructokinase_(PFK)/Pfk_ab_closeup/1'>A/B allosteric site</scene> of subunit A shows contributions from both subunits to the binding of ADP. The conformational changes in going between the R and T states of PFK are illustrated below. | ||
Revision as of 10:52, 3 April 2023
| |||||||||||
Additional Resources
For additional information, see: Carbohydrate Metabolism
References
- ↑ Schirmer T, Evans PR. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990 Jan 11;343(6254):140-5. PMID:2136935 doi:http://dx.doi.org/10.1038/343140a0
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: structure and control. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):53-62. PMID:6115424
- ↑ http://www.nature.com/nature/journal/v327/n6121/abs/327437a0.html
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ PubMed:2136935
- ↑ Campos G, Guixe V, Babul J. Kinetic mechanism of phosphofructokinase-2 from Escherichia coli. A mutant enzyme with a different mechanism. J Biol Chem. 1984 May 25;259(10):6147-52. PMID:6233271
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ PubMed:2136935
- ↑ Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry: Life at the Molecular Level. Hoboken, NJ: Wiley, 2008. Print.
- ↑ Campos G, Guixe V, Babul J. Kinetic mechanism of phosphofructokinase-2 from Escherichia coli. A mutant enzyme with a different mechanism. J Biol Chem. 1984 May 25;259(10):6147-52. PMID:6233271
- ↑ Kimmel JL, Reinhart GD. Reevaluation of the accepted allosteric mechanism of phosphofructokinase from Bacillus stearothermophilus. Proc Natl Acad Sci U S A. 2000 Apr 11;97(8):3844-9. PMID:10759544 doi:10.1073/pnas.050588097
- ↑ Vora S, Corash L, Engel WK, Durham S, Seaman C, Piomelli S. The molecular mechanism of the inherited phosphofructokinase deficiency associated with hemolysis and myopathy. Blood. 1980 Apr;55(4):629-35. PMID:6444532
External Links
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Alexander Berchansky, Judy Voet, Ann Taylor, Jaime Prilusky, David Canner, Eran Hodis, Joel L. Sussman, Zach Westrick