Sandbox Reserved 1777
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
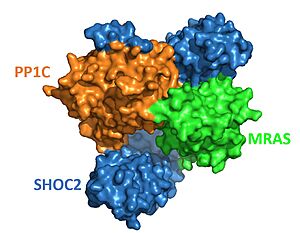
| - | The enzyme requires 3 domains, SHOC-2(blue), PP1C(coral), and MRAS(green), to form the active enzyme (SMP Complex), also known as a holoenzyme (Figure 1)<Ref name='Hauseman'>Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. [https://doi.org/10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1]. </Ref>. | + | The enzyme requires 3 domains, SHOC-2(blue), PP1C(coral), and MRAS(green), to form the active enzyme (SMP Complex), also known as a holoenzyme (Figure 1)<Ref name='Hauseman'>Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. [https://doi.org/10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1]. </Ref>. . The SMP complex was determined via cryo-electron microscopy as well as x-ray diffraction. These studies found that PP1C and MRAS occupy the concave surface of SHOC2, leaving the catalytic site of PP1C and the substrate binding cleft in MRAS exposed. |
==SHOC2== | ==SHOC2== | ||
| + | SHOC2 is a scaffolding protein which acts as a cradle to bind PP1C and MRAS, allowing for the holoenzyme to be functional. The <scene name='95/952706/Shoc2_structure/7'>structure of SHOC2</scene> is a leucine rich repeat (LRR) protein that consists of 20 consecutive <scene name='95/952706/Shoc2_structure/6'>LRR regions</scene>. This motif results in an extended beta sheet on the inner concavity of the protein surface with alpha helices facing outward. This results in a largely hydrophobic core. | ||
==PP1C== | ==PP1C== | ||
| + | Protein phosphatase 1 catalytic subunit is a catalytic domain of a phosphatase enzyme [https://proteopedia.org/wiki/index.php/Protein_phosphatase. PP1], which cleaves a phosphate. <scene name='95/952706/Pp1c_structure/3'>PP1C</scene> is a highly characterized serine/ threonine phosphatase. PP1C is a serine/threonine phosphatase known to be involved in a variety of cellular signalling pathways that control cell growth, division, and metabolism (again cite, see texts). | ||
==MRAS== | ==MRAS== | ||
| + | <scene name='95/952705/Mras_structure/1'>MRAS</scene> is a GTPase protein and is located near (typically just below) the cell membrane. When MRAS binds GTP, it becomes active and triggers the assembly of the active holoenzyme<ref name="Hauseman" />. MRAS is a close relative of the RAS protein and therefore shares most of it's regulatory and effector interactions (CITE, see text from emily lol). A unique function of MRAS is it's ability to form a complex with SHOC2 and PP1, allowing it to have phosphatase activity. | ||
===Switch I and II=== | ===Switch I and II=== | ||
| Line 25: | Line 28: | ||
==Special Interactions== | ==Special Interactions== | ||
| - | + | binds to SHOC2 exclusively through this concaved region <Ref name='Kwan'>Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. [https://doi.org/10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2]. </Ref>, primarily by the descending loop and strands of each LRR domains 2-10. This reaction is stabilized through <scene name='95/952705/Mras_structure/3'>hydrogen bonding</scene>. PP1C <scene name='95/952706/Shoc2_pp1c_interaction/2'>associates</scene> with the ascending loops of the SHOC2 LRR regions, and is further engaged through the N-terminal region containing the <scene name='95/952705/Shoc2_rvxf/1'>RVxF</scene> motif <Ref name= 'Jajian'>Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. [https://aacrjournals.org/cancerres/article/82/12_Supplement/LB029/699443. DOI: 10.1158/1538-7445.AM2022-LB029]. </Ref>. The initial forming of the complex begins with SHOC2:PP1C engagement, then is completed and stabilized by the GTP-loaded MRAS binding <Ref name="Jajian" />. Once MRAS has associated with SHOC2, it <scene name='95/952705/Mras_pp1c_interaction/1'>binds</scene> to PP1C and guides the holoenzyme complex to the cell membrane to begin signaling<ref name="Kwan" />. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
=Mechanism= | =Mechanism= | ||
Revision as of 18:02, 3 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ 1.0 1.1 Bernal Astrain G, Nikolova M, Smith MJ. Functional diversity in the RAS subfamily of small GTPases. Biochem Soc Trans. 2022 Apr 29;50(2):921-933. doi: 10.1042/BST20211166. DOI:10.1042/BST20211166.
- ↑ Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006 Jan;1(1):7-9. DOI:10.1016/S1556-0864(15)31506-9.
- ↑ Li, L., Zhao, G. D., Shi, Z. et. al.The Ras/Raf/MEK/ERK signaling pathway and its role in the occurrence and development of HCC. Oncology letters, 12(5), 3045–3050. DOI:10.3892/ol.2016.5110.
- ↑ 4.0 4.1 Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1.
- ↑ Daniel A. Bonsor, Patrick Alexander, Kelly Snead, Nicole Hartig, Matthew Drew, Simon Messing, Lorenzo I. Finci, Dwight V. Nissley, Frank McCormick, Dominic Esposito, Pablo Rodrigiguez-Viciana, Andrew G. Stephen, Dhirendra K. Simanshu. Structure of the SHOC2–MRAS–PP1C complex provides insights into RAF activation and Noonan syndrome. bioRxiv. 2022.05.10.491335. doi: 10.1101/2022.05.10.491335. DOI:10.1101/2022.05.10.491335.
- ↑ 6.0 6.1 Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2.
- ↑ 7.0 7.1 7.2 Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. DOI: 10.1158/1538-7445.AM2022-LB029.
- ↑ 8.0 8.1 Lavoie, H., Therrien, M. Structural keys unlock RAS–MAPK cellular signalling pathway. Nature 609, 248-249 (2022). doi: 10.1038/d41586-022-02189-7. DOI:10.1038/d41586-022-02189-7.
- ↑ Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J. Structural basis for SHOC2 modulation of RAS signalling. Nature. 2022 Jun 29. pii: 10.1038/s41586-022-04838-3. doi:, 10.1038/s41586-022-04838-3. PMID:35768504 doi:http://dx.doi.org/10.1038/s41586-022-04838-3