Sandbox Reserved 1786
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
=='''Structure'''== | =='''Structure'''== | ||
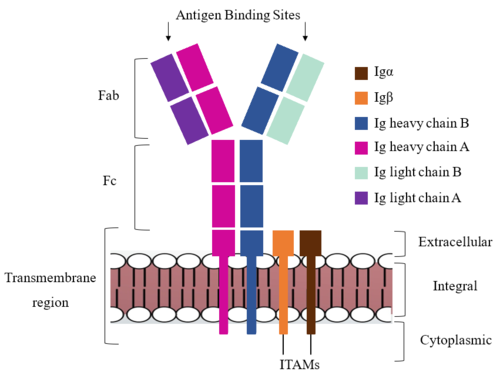
| - | + | The IgM BCR consists of six separate chains (Figure 1) that make up three main domains in the molecule. A depiction of the IgM <scene name='95/952714/Colored_by_domain/3'>colored by domain</scene> shows two heavy and two light chains together form the <b><span class="text-lightblue">Fab region</span></b>, or variable fragment at the top of the molecule where the antigen binding sites are located. The two heavy chains extend below the <b><span class="text-lightblue">Fab region</span></b> through the <b><span class="text-purple">Fc region</span></b> and eventually connect to the Igα/β heterodimer to form the <b><span class="text-orange">transmembrane region</span></b> which anchors the overall complex to the B cell. The overall structure, expression, and function of the IgM BCR has been found to be strongly influenced by the <b><span class="text-orange">transmembrane region</span></b> in which Ig α/β interactions as a heterodimer influence cell surface expression, receptor assembly, and effective signal transduction. In each domain, interactions between individual chains are important to understand the complex as a whole. All future 3D depictions will be <scene name='95/952713/Colored_by_chain/2'>colored by chain</scene> as in this image. | |
| + | [[Image:IgM_structure_overview_diagram.png|500 px|left|thumb|'''Figure 1. IgM BCR Structure Overview.''' Depiction of the IgM BCR expressed on the membrane of a B cell. Includes all major components including the α/β heterodimer, heavy and light chains, antigen binding sites, and the ITAM region for signal transduction.]] | ||
{{Clear}} | {{Clear}} | ||
| Line 17: | Line 18: | ||
The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/11'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively. IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked glycosylation</span></b> (NAGs) on various asparagine residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] remain bound to the alpha and beta subunits until both dimerizations occur; at this point the rest of the BCR complex can be recruited. | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/11'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively. IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked glycosylation</span></b> (NAGs) on various asparagine residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] remain bound to the alpha and beta subunits until both dimerizations occur; at this point the rest of the BCR complex can be recruited. | ||
| - | |||
| - | [[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | ||
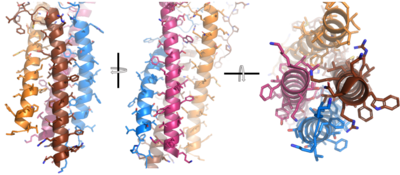
After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The 4 helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The 4 helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. | ||
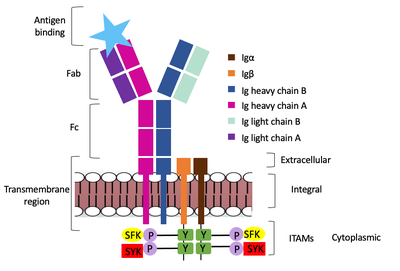
Furthermore, both the Igα and Igβ chains have cytoplasmic tails that extend into the B cell (Figure 1). Each of these tails contain a immuno-receptor tyrosine-based activation motif (ITAM) region to facilitate signal transduction (Figure 4). | Furthermore, both the Igα and Igβ chains have cytoplasmic tails that extend into the B cell (Figure 1). Each of these tails contain a immuno-receptor tyrosine-based activation motif (ITAM) region to facilitate signal transduction (Figure 4). | ||
| + | [[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | ||
{{Clear}} | {{Clear}} | ||
| Line 34: | Line 34: | ||
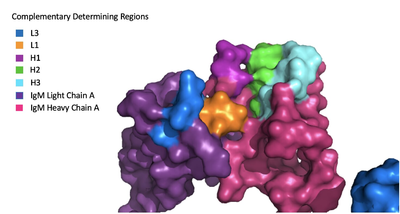
Because the Fab region of IgM is so poorly resolved, a structural comparison to another antibody was performed to approximate where an antigen would bind to the <scene name='95/952713/Variable_region/1'>variable region</scene>. Figure 2 shows the antigen binding motif (located on the Fab region) of an Ig-BCR complex that was engineered to contain the variable region of a neutralizing antibody called VCR01, an antibody that targets the epitope of HIV molecules. It contains areas referred to as complementary-determining regions, or CDRs, which are where the antigen makes contact with the antibody on the Fab domain. Showing them as surface representation allows us to make structural comparisons to the IgM antibody, and to highlight their similarities. The CDRs are similarly placed within the heavy and light chain variable regions between both antibodies. It is speculated that they are structurally similar because the VCR01 antibody can effectively target multiple HIV strains while IgM is the preliminary antibody produced and released during early stages of the immune response, thus it is able to respond in larger concentrations while antibodies that are more specific to the antigen are being produced. | Because the Fab region of IgM is so poorly resolved, a structural comparison to another antibody was performed to approximate where an antigen would bind to the <scene name='95/952713/Variable_region/1'>variable region</scene>. Figure 2 shows the antigen binding motif (located on the Fab region) of an Ig-BCR complex that was engineered to contain the variable region of a neutralizing antibody called VCR01, an antibody that targets the epitope of HIV molecules. It contains areas referred to as complementary-determining regions, or CDRs, which are where the antigen makes contact with the antibody on the Fab domain. Showing them as surface representation allows us to make structural comparisons to the IgM antibody, and to highlight their similarities. The CDRs are similarly placed within the heavy and light chain variable regions between both antibodies. It is speculated that they are structurally similar because the VCR01 antibody can effectively target multiple HIV strains while IgM is the preliminary antibody produced and released during early stages of the immune response, thus it is able to respond in larger concentrations while antibodies that are more specific to the antigen are being produced. | ||
| + | |||
| + | Due to the Fab region of the IgM antibody being poorly resolved, the specific side chain interactions between the heavy and light chains have not been determined. This depiction of the <scene name='95/952713/Heavy-light_chain_interface/1'>heavy-light chain interface</scene> shows how the 4 β-sandwiches fit together; heavy chain A and B of the Fab region form a complex with the rest of the molecule via interactions with the heavy A and B of the Fc region, before continuing down into the intracellular domain to interact with the transmembrane region. The light chains however are only connected to the complex by forming interactions with the heavy chains within the Fab region. Although the specific residues within the Fab region have yet to be identified, it is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. | ||
[[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''']] | [[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''']] | ||
| - | |||
| - | Due to the Fab region of the IgM antibody being poorly resolved, the specific side chain interactions between the heavy and light chains have not been determined. This depiction of the <scene name='95/952713/Heavy-light_chain_interface/1'>heavy-light chain interface</scene> shows how the 4 β-sandwiches fit together; heavy chain A and B of the Fab region form a complex with the rest of the molecule via interactions with the heavy A and B of the Fc region, before continuing down into the intracellular domain to interact with the transmembrane region. The light chains however are only connected to the complex by forming interactions with the heavy chains within the Fab region. Although the specific residues within the Fab region have yet to be identified, it is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. | ||
{{Clear}} | {{Clear}} | ||
Revision as of 00:50, 7 April 2023
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
Student Contributors
Detonyeá Dickson, Allison Goss, Jackson Payton