Sandbox Reserved 1786
From Proteopedia
(Difference between revisions)
| Line 5: | Line 5: | ||
=='''Introduction'''== | =='''Introduction'''== | ||
| - | + | The [https://en.wikipedia.org/wiki/Adaptive_immune_system adaptive immune response] possessed by [https://en.wikipedia.org/wiki/Vertebrate vertebrate] animals owes much of its function to [https://en.wikipedia.org/wiki/B_cell B cells]. These specialized immune cells produce [https://en.wikipedia.org/wiki/Antibody antibodies] and Immunoglobulins (Ig), the membrane bound equivalent to antibodies. B cells can produce a variety of Ig compounds including [https://en.wikipedia.org/wiki/Immunoglobulin_G IgG], [https://en.wikipedia.org/wiki/Immunoglobulin_A IgA], [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE], [https://en.wikipedia.org/wiki/Immunoglobulin_D IgD], and [https://en.wikipedia.org/wiki/Immunoglobulin_M IgM]. These antibodies and Ig compounds bind to specific compounds called [https://en.wikipedia.org/wiki/Antigen antigens]. When an IgM combines with a [https://en.wikipedia.org/wiki/B-cell_receptor B cell receptor] (BCR) it can then send a signal in the form of a conformational change through the B cell membrane to stimulate the production of more antibodies that recognize that antigen. (Su and Ma) | |
| - | The structure of IgM was determined using [https:// | + | The structure of the IgM BCR complex was determined by two research groups using [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo EM]. They also determined the structure of IgG. (Su and Ma) |
=='''Structure'''== | =='''Structure'''== | ||
| Line 33: | Line 33: | ||
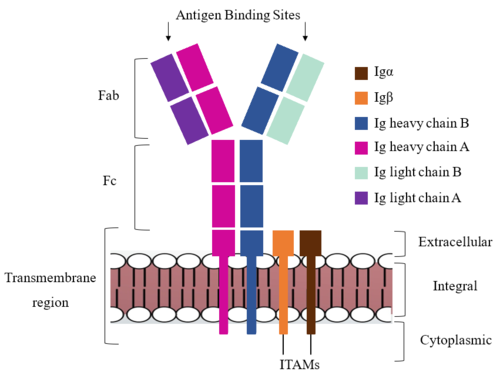
===Fab Region=== | ===Fab Region=== | ||
| - | Because the Fab region of IgM is | + | The Fab region of the antibody is where antigen recognition occurs upon binding. On each arm is one heavy and one light chain, both containing domains identical to their respective counterparts. Repeats of β-sandwiches form the constant and variable domains (blue link) within the Fab region as antigen recognition occurs at the variable domain while the constant domain connects it to the rest of the IgM complex. Because the Fab region of IgM is poorly resolved, a structural analysis of an HIV neutralizing antibody called VCR01 was performed to approximate where an antigen would bind to at the <scene name='95/952713/Variable_region/1'>variable region</scene>. |
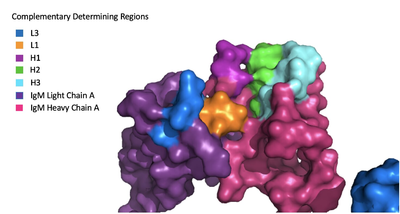
| + | The IgM-BCR contains areas referred to as complementary-determining regions (blue link)(CDRs), which are where the antigen makes contact with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. | ||
| - | Due to the Fab region | + | Due to the poor resolution of the Fab region, specific side chain interactions between the heavy and light chains have not been determined. It is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. The <scene name='95/952713/Heavy-light_chain_interface/1'>heavy-light chain interface</scene> shows how the four heavy and light chain β-sandwiches fit together. The Fab region heavy chains attach to the Fc region heavy chains via the Hinge region (blue link), before continuing down into the intracellular domain to interact with the Igα/Igβ subunits. The light chains however are only connected to the heavy chains within the Fab region, thus have no contact with the subsequent domains. |
[[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''']] | [[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''']] | ||
| Line 42: | Line 43: | ||
=='''Signal Transduction'''== | =='''Signal Transduction'''== | ||
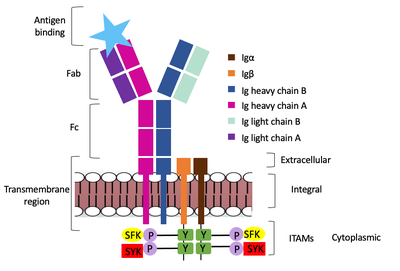
| - | The diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown but it is speculated to operate under what is known as the conserved assembly mechanism. This means that upon antigen binding, BCRs on the surface of the cell begin to cluster to cause the phosphorylation of the immunoreceptor tyrosine-based activation motifs located in Igα and Igβ. In its “off” state, the constant region 4 of heavy chain B overlaps the extracellular components of Igα and Igβ. As the antigen binds, it induces a conformational change to release the overlap and allow for clustering about the BCR. Now, in its “on” state the phosphorylation of the ITAM region (observed here as the conserved tyrosine residues are phosphorylated) within the intracellular tails of Igα and Igβ drives downstream kinase activity to continue to process of signal cascading. | + | The diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown but it is speculated to operate under what is known as the conserved assembly mechanism (blue link). This means that upon antigen binding, BCRs on the surface of the cell begin to cluster to cause the phosphorylation of the immunoreceptor tyrosine-based activation motifs located in Igα and Igβ. In its “off” state, the constant region 4 of heavy chain B overlaps the extracellular components of Igα and Igβ. As the antigen binds, it induces a conformational change to release the overlap and allow for clustering about the BCR. Now, in its “on” state the phosphorylation of the ITAM region (blue link) (observed here as the conserved tyrosine residues are phosphorylated) within the intracellular tails of Igα and Igβ drives downstream kinase activity to continue to process of signal cascading (blue link). |
[[Image:Signal_transduction-2.png|400 px|left|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''']] | [[Image:Signal_transduction-2.png|400 px|left|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''']] | ||
Revision as of 13:04, 7 April 2023
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
Student Contributors
Detonyeá Dickson, Allison Goss, Jackson Payton