Sandbox Reserved 1786
From Proteopedia
(Difference between revisions)
| Line 19: | Line 19: | ||
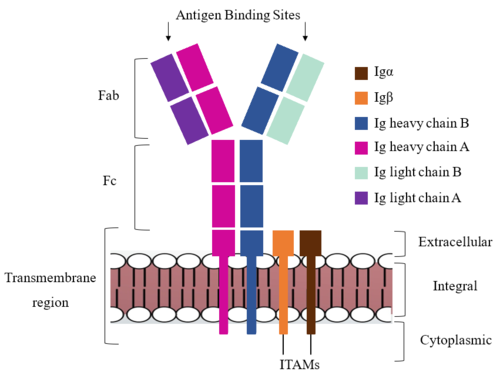
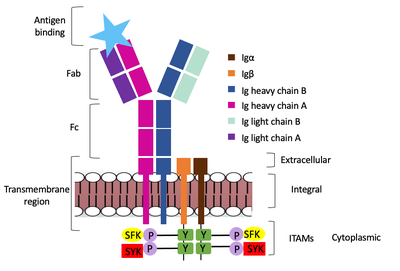
The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/12'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively (Figure 1). IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. (Tolar citation) The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked asparagine glycosylation</span></b> (NAGs) on various residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] are typically bound to the alpha and beta subunits until dimerization occurs; at this point the rest of the BCR complex can be recruited. (Dylke citation) | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/12'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively (Figure 1). IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. (Tolar citation) The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked asparagine glycosylation</span></b> (NAGs) on various residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] are typically bound to the alpha and beta subunits until dimerization occurs; at this point the rest of the BCR complex can be recruited. (Dylke citation) | ||
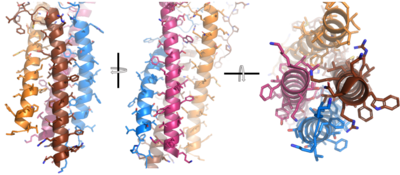
| - | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. (Tolar citation) The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The | + | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. (Tolar citation) The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The four helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. (Dylke citation) |
[[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | [[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | ||
| Line 30: | Line 30: | ||
===Fc Region=== | ===Fc Region=== | ||
| - | The constant region of IgM is made up of the two <scene name='95/952715/Heavy_chain/1'>heavy chains</scene>. These heavy chains form a bridge to connect the Fab fragment, or variable region, to the transmembrane region. They also act as a wire to allow the variable region to send a cellular signal through to the intermembrane region once an antigen has been bound. | + | The constant region of IgM is made up of the two <scene name='95/952715/Heavy_chain/1'>heavy chains</scene> (Figure 1). These heavy chains form a bridge to connect the Fab fragment, or variable region, to the transmembrane region. They also act as a wire to allow the variable region to send a cellular signal through to the intermembrane region once an antigen has been bound. |
Interactions between the <scene name='95/952715/Alpha_beta_heavy/2'>Igα, Igβ, and Heavy chains</scene> help to stabilize and hold the complex together in the extracellular portion of the transmembrane region. | Interactions between the <scene name='95/952715/Alpha_beta_heavy/2'>Igα, Igβ, and Heavy chains</scene> help to stabilize and hold the complex together in the extracellular portion of the transmembrane region. | ||
| Line 40: | Line 40: | ||
===Fab Region=== | ===Fab Region=== | ||
| - | The Fab region of the antibody is where antigen recognition occurs upon binding. On each arm is one heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and one light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chain, both containing domains identical to their respective counterparts. Repeats of β-sandwiches form the [https://en.wikipedia.org/wiki/Antibody constant and variable domains] within the Fab region as antigen recognition occurs at the variable domain while the constant domain connects it to the rest of the IgM complex. Because the Fab region of IgM is poorly resolved, a structural analysis of an HIV neutralizing antibody called VCR01 was performed to approximate where an antigen would bind to at the <scene name='95/952713/Variable_region/1'>variable region</scene>. | + | The Fab region of the antibody is where antigen recognition occurs upon binding (Figure 1). On each arm is one heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and one light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chain, both containing domains identical to their respective counterparts. Repeats of β-sandwiches form the [https://en.wikipedia.org/wiki/Antibody constant and variable domains] within the Fab region as antigen recognition occurs at the variable domain while the constant domain connects it to the rest of the IgM complex. Because the Fab region of IgM is poorly resolved, a structural analysis of an HIV neutralizing antibody called VCR01 was performed to approximate where an antigen would bind to at the <scene name='95/952713/Variable_region/1'>variable region</scene>. |
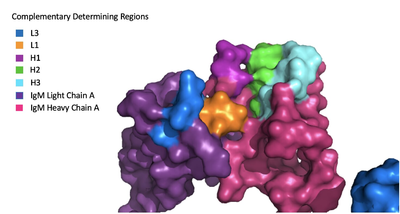
The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes contact with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. | The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes contact with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. | ||
Revision as of 15:39, 7 April 2023
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
Student Contributors
Detonyeá Dickson, Allison Goss, Jackson Payton