Sandbox Reserved 1786
From Proteopedia
(Difference between revisions)
| Line 11: | Line 11: | ||
=='''Structure'''== | =='''Structure'''== | ||
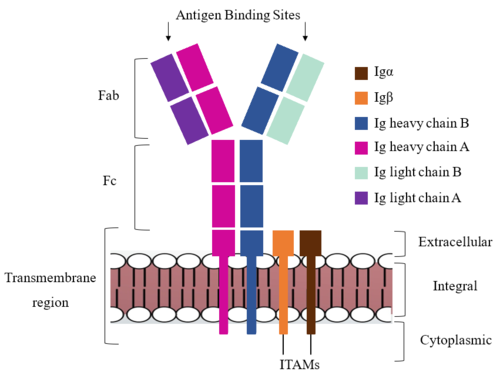
| - | The IgM BCR consists of six separate chains (Figure 1) that make up three main domains in the molecule. A depiction of the IgM <scene name='95/952714/Colored_by_domain/3'>colored by domain</scene> shows two heavy and two light chains that together form the <b><span class="text-cyan">Fab region</span></b>, or variable fragment at the top of the molecule where the antigen binding sites are located. The two heavy chains extend below the <b><span class="text-cyan">Fab region</span></b> through the <b><span class="text-purple">Fc region</span></b> and eventually connect to the Igα/β heterodimer to form the <b><span class="text-orange">transmembrane region</span></b> which anchors the overall complex to the B cell. The overall structure, expression, and function of the IgM BCR has been found to be strongly influenced by the <b><span class="text-orange">transmembrane region</span></b> in which Ig α/β interactions as a heterodimer influence cell surface expression, receptor assembly, and effective signal transduction (Tolar and Dylke citation). In each domain, interactions between individual chains are important to understand the complex as a whole. All future 3D depictions will be <scene name='95/952714/Colored_by_chain/8'>colored by chain</scene> as in Figure 1. | + | The IgM BCR consists of six separate chains (Figure 1) that make up three main domains in the molecule. A depiction of the IgM <scene name='95/952714/Colored_by_domain/3'>colored by domain</scene> shows two heavy and two light chains that together form the <b><span class="text-cyan">Fab region</span></b>, or variable fragment at the top of the molecule where the antigen binding sites are located. The two heavy chains extend below the <b><span class="text-cyan">Fab region</span></b> through the <b><span class="text-purple">Fc region</span></b> and eventually connect to the Igα/β heterodimer to form the <b><span class="text-orange">transmembrane region</span></b> which anchors the overall complex to the B cell. The overall structure, expression, and function of the IgM BCR has been found to be strongly influenced by the <b><span class="text-orange">transmembrane region</span></b> in which Ig α/β interactions as a heterodimer influence cell surface expression, receptor assembly, and effective signal transduction. <ref name="Dylke">PMID:17675166</ref> (Tolar and Dylke citation). In each domain, interactions between individual chains are important to understand the complex as a whole. All future 3D depictions will be <scene name='95/952714/Colored_by_chain/8'>colored by chain</scene> as in Figure 1. |
[[Image:IgM_structure_overview_diagram.png|500 px|left|thumb|'''Figure 1. IgM BCR Structure Overview.''' Depiction of the IgM BCR expressed on the membrane of a B cell. Includes all major components including the α/β heterodimer, heavy and light chains, antigen binding sites, and the ITAM region for signal transduction.]] | [[Image:IgM_structure_overview_diagram.png|500 px|left|thumb|'''Figure 1. IgM BCR Structure Overview.''' Depiction of the IgM BCR expressed on the membrane of a B cell. Includes all major components including the α/β heterodimer, heavy and light chains, antigen binding sites, and the ITAM region for signal transduction.]] | ||
{{Clear}} | {{Clear}} | ||
| Line 17: | Line 17: | ||
===Transmembrane Region=== | ===Transmembrane Region=== | ||
| - | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/12'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively (Figure 1). IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. (Tolar citation) The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked asparagine glycosylation</span></b> (NAGs) on various residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] are typically bound to the alpha and beta subunits until dimerization occurs; at this point the rest of the BCR complex can be recruited. (Dylke citation) | + | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/12'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively (Figure 1). IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. (Tolar citation) The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked asparagine glycosylation</span></b> (NAGs) on various residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] are typically bound to the alpha and beta subunits until dimerization occurs; at this point the rest of the BCR complex can be recruited. <ref name="Dylke">PMID:17675166</ref> (Dylke citation) |
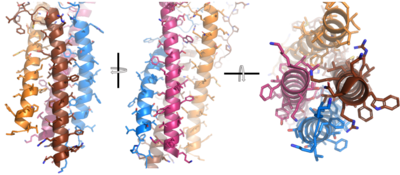
| - | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. (tolar) The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The four helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. (Dylke citation) | + | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. (tolar) The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The four helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. <ref name="Dylke">PMID:17675166</ref> (Dylke citation) |
[[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | [[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | ||
Revision as of 16:31, 7 April 2023
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
- ↑ 1.0 1.1 Su Q, Chen M, Shi Y, Zhang X, Huang G, Huang B, Liu D, Liu Z, Shi Y. Cryo-EM structure of the human IgM B cell receptor. Science. 2022 Aug 19;377(6608):875-880. doi: 10.1126/science.abo3923. Epub 2022, Aug 18. PMID:35981043 doi:http://dx.doi.org/10.1126/science.abo3923
- ↑ 2.0 2.1 2.2 2.3 Ma X, Zhu Y, Dong, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. doi: 10.1126/science.abo3828. Epub 2022, Aug 18. PMID:35981028 doi:http://dx.doi.org/10.1126/science.abo3828
- ↑ 3.0 3.1 3.2 Dylke J, Lopes J, Dang-Lawson M, Machtaler S, Matsuuchi L. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR). Immunol Lett. 2007 Sep 15;112(1):47-57. PMID:17675166 doi:10.1016/j.imlet.2007.06.005
Student Contributors
DeTonyeá Dickson, Allison Goss, Jackson Payton