Introduction

"Fig. 1 Image of Taurocholic acid a crystalline bile acid"
Sodium Taurocholate Co-Transporting Polypeptide, or NTCP, is a membrane transporter protein that is found in the plasma membrane of liver cells, or hepatocytes. NTCP's primary function is the transportation of taurocholates, or bile salts, (Fig. 1) into the liver and out of the liver to the small intestine. [1] Bile salts play various roles in metabolism and digestion, but their main function is the emulsification of lipid droplets into smaller fragments. This enables lipases to break down the droplets into their monomers, or triglycerides which are then able to be digested. NTCP is part of the solute carrier superfamily, more specifically SLC10. NTCP is the founding member of the SLC10 family, first discovered in rat hepatocytes in 1978. [2] NTCP has a key role in Enterohepatic circulation or bile salt recycling, and its unique ability to transport other solutes lends it therapeutic potential for lowering cholesterol and treating liver disease.
NTCP also serves as a binding site for hepatitis B virus and hepatitis D virus. [3] Future studies into HBV binding mechanism can help understand infection pathways and the development of viral inhibitors.
Structure
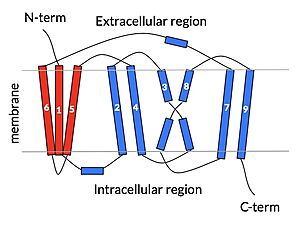
Figure 2: cartoon depiction of NTCP topology Overview
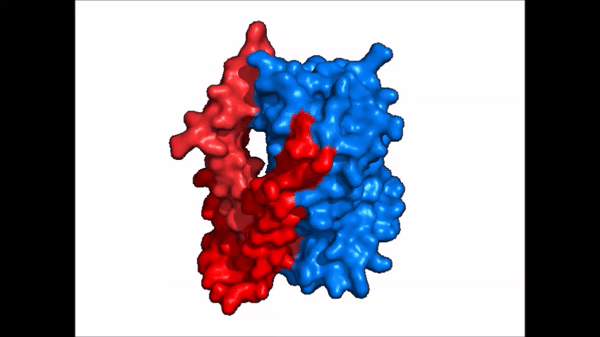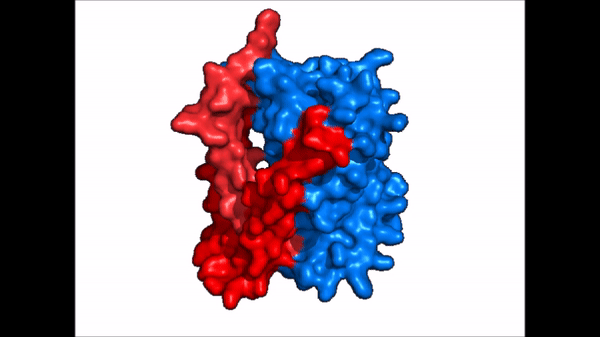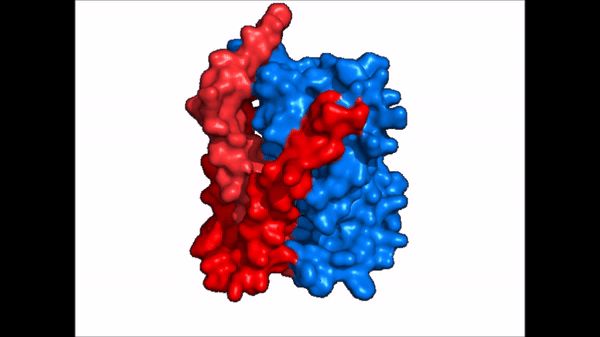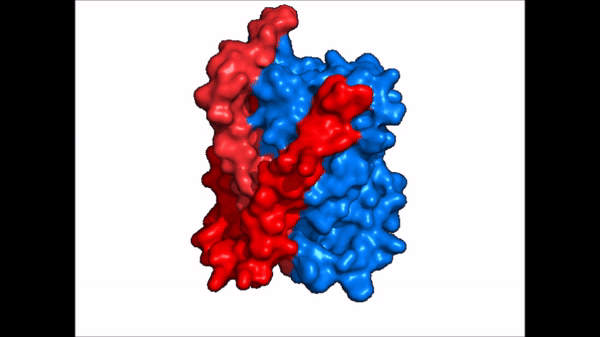
NTCP is one continuous polypeptide chain consisting of a total of . The N-terminus of the polypeptide chain is found on the extracellular region of the plasma membrane while the C-terminus is located on the intracellular region. There are within the quaternary structure of NTCP: a core domain and a panel domain both being a part of the same polypeptide chain (Fig. 2). The (blue) includes 6 transmembrane α helices (TM2-4 and TM7-9) and demonstrates two-fold pseudosymmetry. The (red) consists of 3 transmembrane α helices (TM1 and TM5-6) and does not display symmetry. Within the core domain, there is a unique crossover between TM-3 and TM-8 that is known as the . This motif is important because this is where the transporter's substrate binding site is located, and within this motif lies essential residues that aid in the conformational change that NTCP undergoes. The core and panel domains are connected by both extracellular and intracellular that are separate from the nine transmembrane α helices.
Binding Sites
Sodium
NTCP, among others in the SLC10 family, have . Many polar and negatively charged residues are characteristic of these active sites. The high level of conservation among sodium binding placement and interacting residues suggests sodium binding is coupled to bile salt transport. Additional mutations in the X-motif near sodium binding sites have shown that bile salt transport function is lost. This suggests sodium binding impacts bile salt binding.
[4] It is understood that sodium binding and release facilitates changes from open-pore to inward-facing states of NTCP. The inward-facing state is favored in the absence of sodium ions, while the open-pore state is favored in the presence of sodium ions. This also allows for sodium concentrations to regulate the uptake of taurocholates. When intracellular sodium levels are higher, the open-pore state is favored allowing for the diffusion of taurocholates. However, when extracellular sodium levels are high, the inward-facing state is favored preventing diffusion of taurocholates. [4]
Bile Salts
The is also characteristic of NTCP. The pore surface remains Hydrophobic, while lining of the open pore state is largely Polar. However, in the "inward-facing conformation" the polar pore residues are inaccessible. When the pore is closed only the surface hydrophobic residues are observed. As the pore opens up inner polar residues become accessible allowing for the binding of substrates. The pattern of hydrophobic and polar residues within the pore is believed to follow similar amphipathic patterns within taurocholate and other NTCP substrates, such as steroids and thyroid hormones. [5] Thus the channel provides specificity while preventing leakage of other substrates. When observing the relevant it is shown that some residues form Van der Waals interactions while others will form dipole-dipole or ionic interactions with bile salt substrates. The core domain appears to contribute most of the polar domains, while the panel domain contributes more hydrophobic residues.
Conformational Change
| | 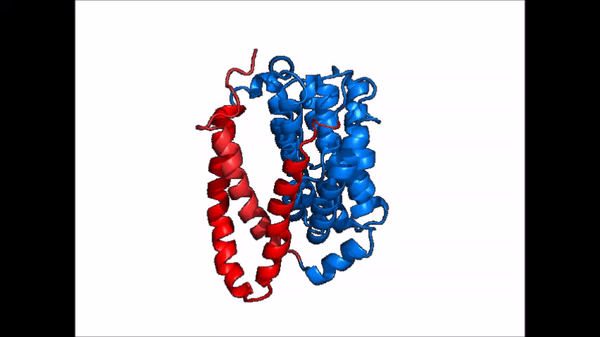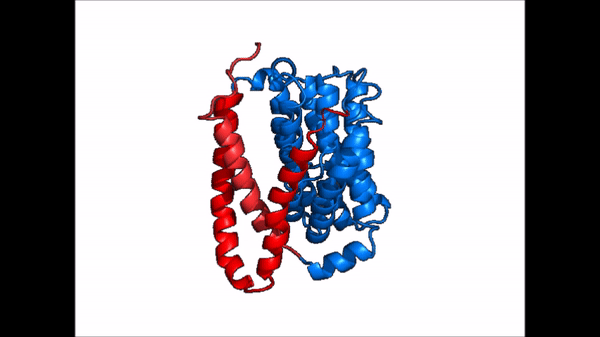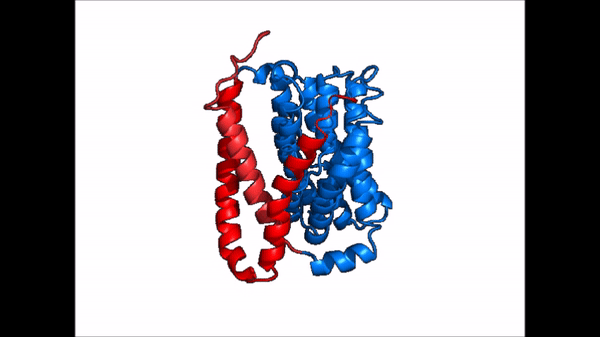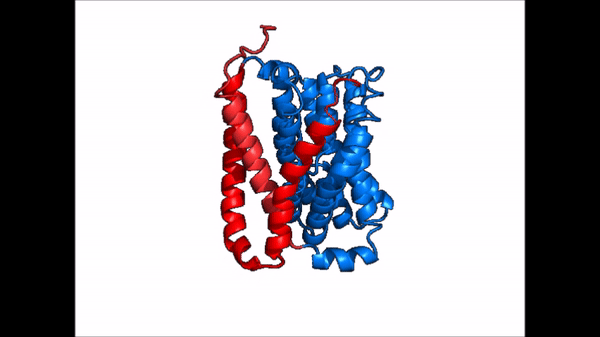 |
| Cartoon representation of NTCP conformational change. |
| |  |
| Cartoon representation of NTCP conformational change. |
NTCP exists in two different conformations; the and the . In order to transport bile salts across the plasma membrane of hepocytes, NTCP must undergo the conformational change from inward facing to open pore. This movement consists of the core and panel domains both rotating 20° and the panel domain moving 5 Å away from the core domain, which remains relatively rigid. This conformational change reveals the two sodium ion binding sites as well as the amphipathic pore in the membrane that bile salts can pass through. This movement of the panel domain is facilitated by located in the connector helices between the panel and core domains. These residues act as hinges that assist in the movement of the panel domain away from the core domain.
Bile Salt Transport
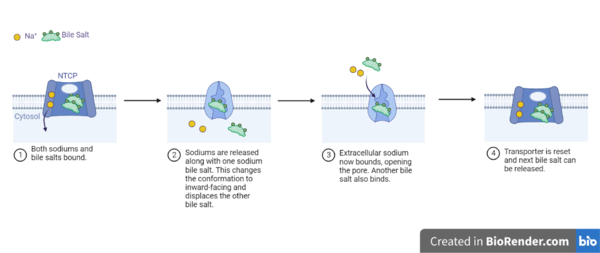
Figure 5: Diagram of Proposed Bile Salt Transport Process A proposed pathway for NTCP bile salt transport suggests that both sodium ions are translocated with the transport of one bile salt.[6] Initally all then both sodium ions are released along with the inner bile salt into the cytoplasm (Fig. 5). The however in the pore, likely helping to prevent leakage. [6] The into the inner bile salt placement by the movement of sodium ions that facilitates the conformational change to the inward-facing, pore inaccessible conformation (Fig. 5). [6] It utilizes an elevator-alternating mechanism where one domain (core) does most of the translocation, and the other domain (panel) remains stationary. [7] Sodium ions then bind to NTCP, favoring the open-pore state and also allowing for the binding of another outer bile salt (Fig 5). The and the process can then start again releasing the next inner bile salt with the translocation of the sodium ions into the cytoplasm.
HBV Binding and Infection
NTCP is the only entry receptor into the liver for HBV. [7] The myristolated PreS1 domain of HBV binds to NTCP through a containing residues 157-165 on the open pore surface. [7] These residues form part of the tunnel resulting in HBV binding and bile salt transport directly competing and interfering with one another. [7] Another hydrophobic patch consisting of residues 84-87 found on the N-terminus of NTCP does not overlap with bile salt binding and may be used for the development of antivirals that don't inhibit bile uptake [3]. Other minor variations within NTCP provide species specificity for HBV or virus resistance, such as mutant S267F found in East Asia. [3]
The exact mechanism by which NTCP mediates viral internalization is still yet to be determined; however, current studies speculate it works through endocytosis. [8] Once HBV is bound the NTCP/HBV complex is taken into the cell where viral contents are dumped into the cytoplasm to then begin viral replication. It is currently unknown whether HBV also interacts with other receptors or host cell factors, but NTCP alone is not sufficient for infection. [8]
Medical Relevancy
This is a sample scene created with SAT to by Group, and another to make of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes.





