This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Sandbox Reserved 1785
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
='''Human B-cell Antigen Receptor: IgM BCR'''= | ='''Human B-cell Antigen Receptor: IgM BCR'''= | ||
| - | <StructureSection load=' | + | <StructureSection load='7xq8' size='350' side='right' caption='IgM B-Cell Receptor (PDB: 7xq8)' scene='95/952714/Colored_by_chain/6'> |
=='''Introduction'''== | =='''Introduction'''== | ||
| - | The [https://en.wikipedia.org/wiki/Adaptive_immune_system adaptive immune response] possessed by [https://en.wikipedia.org/wiki/Vertebrate vertebrate] animals owes much of its function to [https://en.wikipedia.org/wiki/B_cell B cells]. These specialized immune cells produce [https://en.wikipedia.org/wiki/Antibody antibodies] and Immunoglobulins (Ig), the membrane bound equivalent to antibodies. B cells can produce a variety of Ig compounds including [https://en.wikipedia.org/wiki/Immunoglobulin_G IgG], [https://en.wikipedia.org/wiki/Immunoglobulin_A IgA], [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE], [https://en.wikipedia.org/wiki/Immunoglobulin_D IgD], and [https://en.wikipedia.org/wiki/Immunoglobulin_M IgM]. These antibodies and Ig compounds bind to specific compounds called [https://en.wikipedia.org/wiki/Antigen antigens]. When an IgM combines with a [https://en.wikipedia.org/wiki/B-cell_receptor B cell receptor] (BCR) it can then send a signal in the form of a conformational change through the B cell membrane to stimulate the production of more antibodies that recognize that antigen. ( | + | The [https://en.wikipedia.org/wiki/Adaptive_immune_system adaptive immune response] possessed by [https://en.wikipedia.org/wiki/Vertebrate vertebrate] animals owes much of its function to [https://en.wikipedia.org/wiki/B_cell B cells]. These specialized immune cells produce [https://en.wikipedia.org/wiki/Antibody antibodies] and Immunoglobulins (Ig), the membrane bound equivalent to antibodies. B cells can produce a variety of Ig compounds including [https://en.wikipedia.org/wiki/Immunoglobulin_G IgG], [https://en.wikipedia.org/wiki/Immunoglobulin_A IgA], [https://en.wikipedia.org/wiki/Immunoglobulin_E IgE], [https://en.wikipedia.org/wiki/Immunoglobulin_D IgD], and [https://en.wikipedia.org/wiki/Immunoglobulin_M IgM]. These antibodies and Ig compounds bind to specific compounds called [https://en.wikipedia.org/wiki/Antigen antigens]. When an IgM combines with a [https://en.wikipedia.org/wiki/B-cell_receptor B cell receptor] (BCR) it can then send a signal in the form of a conformational change through the B cell membrane to stimulate the production of more antibodies that recognize that antigen. <ref name="Sathe">Sathe A, Cusick JK. Biochemistry, Immunoglobulin M. 2022 Dec 19. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan–. PMID: 32310455. https://pubmed.ncbi.nlm.nih.gov/32310455/</ref> |
| - | The structure of the IgM BCR complex was determined by two research groups using [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo EM]. They also determined the structure of IgG. | + | The structure of the IgM BCR complex was determined by two research groups using [https://en.wikipedia.org/wiki/Cryogenic_electron_microscopy Cryo EM]. They also determined the structure of IgG. <ref name="Su">PMID:35981043</ref>, <ref name="Ma">PMID:35981028</ref> |
=='''Structure'''== | =='''Structure'''== | ||
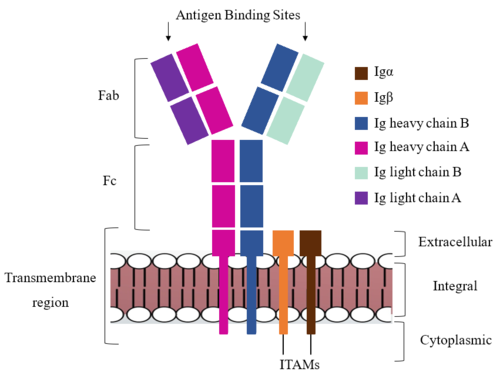
| - | The IgM BCR consists of six separate chains (Figure 1) that make up three main domains in the molecule. A depiction of the IgM <scene name='95/952714/Colored_by_domain/3'>colored by domain</scene> shows two heavy and two light chains together form the <b><span class="text- | + | [[Image:IgM_structure_overview_diagram.png|500 px|left|thumb|'''Figure 1. IgM BCR Structure Overview.''' Depiction of the IgM BCR expressed on the membrane of a B cell. Includes all major components including the α/β heterodimer, heavy and light chains, antigen binding sites, and the ITAM region for signal transduction.]]The IgM BCR consists of six separate chains (Figure 1) that make up three main domains in the molecule. A depiction of the IgM <scene name='95/952714/Colored_by_domain/3'>colored by domain</scene> shows two heavy and two light chains that together form the <b><span class="text-cyan">Fab region</span></b>, or variable fragment at the top of the molecule where the antigen binding sites are located. The two heavy chains extend below the <b><span class="text-cyan">Fab region</span></b> through the <b><span class="text-purple">Fc region</span></b> and eventually connect to the Igα/β heterodimer to form the <b><span class="text-orange">transmembrane region</span></b> which anchors the overall complex to the B cell. The overall structure, expression, and function of the IgM BCR has been found to be strongly influenced by the <b><span class="text-orange">transmembrane region</span></b> in which Ig α/β interactions as a heterodimer influence cell surface expression, receptor assembly, and effective signal transduction. <ref name="Tolar">Tolar P, Pierce SK. Unveiling the B cell receptor structure. Science. 2022 Aug 19;377(6608):819-820. doi: 10.1126/science.add8065. Epub 2022 Aug 18.[http://dx.doi.org/10.1126/science.add8065 DOI:10.1126/science.add8065</ref>, <ref name="Dylke">Dylke J, Lopes J, Dang-Lawson M, Machtaler S, Matsuuchi L. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR). Immunol Lett. 2007 Sep 15;112(1):47-57. doi: 10.1016/j.imlet.2007.06.005. Epub 2007 Jul 23. [http://dx.doi.org/10.1016/j.imlet.2007.06.005 DOI:10.1016/j.imlet.2007.06.005</ref>. In each domain, interactions between individual chains are important to understand the complex as a whole. All future 3D depictions will be <scene name='95/952714/Colored_by_chain/8'>colored by chain</scene> as in Figure 1. |
| - | + | ||
{{Clear}} | {{Clear}} | ||
===Transmembrane Region=== | ===Transmembrane Region=== | ||
| - | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/ | + | The IgM BCR is anchored to [https://en.wikipedia.org/wiki/B_cell B-cell] membranes through the <scene name='95/952714/Integral_region/13'>transmembrane region</scene> which is broken up into both extracellular and integral domains which sit on top of or span through the membrane, respectively (Figure 1). IgM BCR assembly requires dimerization of the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> subunits which embed within the B-cell membrane. <ref name="Tolar"/> The <scene name='95/952714/Ig_alpha_beta/5'>Igα and Igβ heterodimer</scene> dimerizes within the extracellular region with a <scene name='95/952714/Extracellular_disulfide_bridge/6'>disulfide bridge</scene>. Additional dimerization is believed to occur within the integral region via a hydrogen bond; the involved residues and interaction have not been confirmed. Although the mechanism of disulfide bridge formation is still unknown, it is believed that <scene name='95/952714/Extracellular_glycosylation/2'>extracellular glycosylation</scene> via <b><span class="text-lightgreen">N-linked asparagine glycosylation</span></b> (NAGs) on various residues in the extracellular region of both the <b><span class="text-brown">Igα</span></b> and and <b><span class="text-orange">Igβ</span></b> chains help facilitate this process. [https://en.wikipedia.org/wiki/Chaperone_(protein) Chaperone proteins] are typically bound to the alpha and beta subunits until dimerization occurs; at this point the rest of the BCR complex can be recruited. <ref name="Dylke"/> |
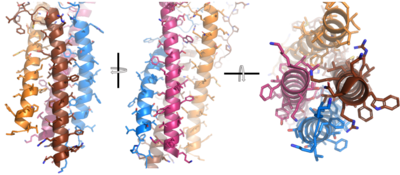
| - | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The | + | After <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> dimerization, the transmembrane helices of the heavy chains can embed within the B-cell membrane. <ref name="Tolar"/> The side chains of this <scene name='95/952714/Integral_helices_2/2'>4-pass integral helix structure</scene> are primarily hydrophobic side chains that allow for interactions with the hydrophobic tails in the [https://en.wikipedia.org/wiki/Lipid_bilayer phospholipid bilayer]. The four helices (Figure 2) are primarily held together through hydrophobic interactions; however, a a few polar residues are included on the interior of the helix structure which interact with a few polar residues on the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains. <ref name="Dylke"/> |
| - | Within the transmembrane region, heavy chains A and B associate asymmetrically to facilitate intracellular signaling cascades. The <scene name='95/952713/Trans_heavy/9'>heavy chain interface</scene> allows them to pack together via [https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals] contacts, but there are also prominent hydrogen bonds between each chain. More specifically, the hydroxyl group from Ser584 on heavy chain A donates a hydrogen bond to Ser584 and to Ser588 on heavy chain B. This creates a [https://en.wikipedia.org/wiki/Hydrogen_bond bifurcated hydrogen bond], essentially forming a “fork” between the two chains to help stabilize them and maintain the transmission of the signal once the cell is activated. Because transmembrane Ig molecules cannot efficiently initiate the signal cascade, they must associate with the Igα and Igβ proteins w/in the BCR (cite). | ||
| - | |||
| - | Furthermore, both the Igα and Igβ chains have cytoplasmic tails that extend into the B cell (Figure 1). Each of these tails contain an immuno-receptor tyrosine-based activation motif (ITAM) region to facilitate signal transduction (Figure 4). | ||
[[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | [[Image:Integral_helix_figure.png|400 px|left|thumb|'''Figure 2. 4-pass integral helix.''' Pymol image of the integral helices in IgM BCR (PDB:7xq8) rotated on the x and y axes. Side chains are shown as sticks. Brown=Ig alpha, orange=Ig beta, pink=heavy chain A, blue=heavy chain B.]] | ||
{{Clear}} | {{Clear}} | ||
| + | |||
| + | Within the transmembrane region, '''{{Font color|violet|heavy chain A}}''' and <b><span class="text-blue">heavy chain B</span></b> associate (Figure 1) asymmetrically to facilitate intracellular signaling cascades. The <scene name='95/952713/Trans_heavy/9'>transmembrane heavy chain interface</scene> allows them to pack together via [https://en.wikipedia.org/wiki/Van_der_Waals_force Van der Waals] contacts, but there are also prominent hydrogen bonds between each chain. More specifically, the hydroxyl group from Ser584 on '''{{Font color|violet|heavy chain A}}''' donates a hydrogen bond to Ser584 and to Ser588 on <b><span class="text-blue">heavy chain B</span></b>. This creates a [https://en.wikipedia.org/wiki/Hydrogen_bond bifurcated hydrogen bond], essentially forming a “fork” between the two chains to help stabilize them and maintain the transmission of the signal once the cell is activated. Because transmembrane Ig molecules cannot efficiently initiate the signal cascade, they must associate with the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains within the BCR. <ref name="Su">PMID:35981043</ref> | ||
| + | |||
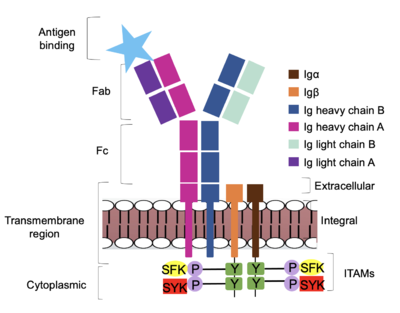
| + | Furthermore, both the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> chains have cytoplasmic tails that extend into the B cell (Figure 1). Each of these tails contain an [https://en.wikipedia.org/wiki/Immunoreceptor_tyrosine-based_activation_motif ITAM region] to facilitate signal transduction (Figure 4). <ref name="Ma">PMID:35981028</ref> | ||
===Fc Region=== | ===Fc Region=== | ||
| - | The constant region of IgM is made up of the two <scene name='95/ | + | The constant region of IgM is made up of the two <scene name='95/952714/Heavy_chain/1'>heavy chains</scene>. These heavy chains form a bridge connecting the FAB region or variable region to the transmembrane region (Figure 1). They also act as a wire that the variable region can send a signal through to the transmembrane region as a mechanical change. |
| - | <scene name='95/ | + | <scene name='95/952714/Extracellular_transmembrane_v2/1'>Extracellular transmembrane region interactions</scene> help hold the heavy chains and <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> chains together in the extracellular portion of the transmembrane region. |
| - | Because a conformational change occurs throughout the entirety of the IgM-BCR complex, the Fc region must be able to tolerate the contortion of the molecule as the antigen binds. In constant region two, which is located at the start of the Fc region, heavy | + | Because a conformational change occurs throughout the entirety of the IgM-BCR complex, the Fc region must be able to tolerate the contortion of the molecule as the antigen binds. In constant region two, which is located at the start of the Fc region, '''{{Font color|violet|heavy chain A}}''' and <b><span class="text-blue">heavy chain B</span></b> make a <scene name='95/952713/Disulfides/5'>disulfide bridge</scene> to stabilize the IgM-BCR and drive downstream signaling. |
| - | To maximize the Fc region’s signal transduction efficiency and Van der Waals contacts, constant region two of heavy chain A makes an asymmetrical association with constant region three of heavy chain B to create a <scene name='95/952713/Trans_heavy/7'>heavy chain interface</scene>. More specifically, Arg243 and Arg251 residues from heavy chain A donate three hydrogen bonds to Leu433, Thr431, and Asp376 residues on heavy chain B. Furthermore, Leu313 of heavy chain A accepts a hydrogen bond from Thr429 on heavy chain B | + | To maximize the Fc region’s signal transduction efficiency and Van der Waals contacts, constant region two of '''{{Font color|violet|heavy chain A}}''' makes an asymmetrical association with constant region three of <b><span class="text-blue">heavy chain B</span></b> to create a <scene name='95/952713/Trans_heavy/7'>heavy chain interface</scene>. More specifically, Arg243 and Arg251 residues from '''{{Font color|violet|heavy chain A}}''' donate three hydrogen bonds to Leu433, Thr431, and Asp376 residues on <b><span class="text-blue">heavy chain B</span></b>. Furthermore, Leu313 of heavy chain A accepts a hydrogen bond from Thr429 on heavy chain B. <ref name="Ma">PMID:35981028</ref> |
===Fab Region=== | ===Fab Region=== | ||
| - | The Fab region of the antibody is where antigen recognition occurs upon binding. On each arm is one heavy and one light chain, both containing domains identical to their respective counterparts. Repeats of β-sandwiches form the [https://en.wikipedia.org/wiki/Antibody constant and variable domains] within the Fab region as antigen recognition occurs at the variable domain while the constant domain connects it to the rest of the IgM complex. Because the Fab region of IgM is poorly resolved, a structural analysis of an HIV neutralizing antibody called VCR01 was performed to approximate where an antigen would bind to at the <scene name='95/ | + | The Fab region of the antibody is where antigen recognition occurs upon binding (Figure 1). On each arm is one heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and one light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chain, both containing domains identical to their respective counterparts. Repeats of β-sandwiches form the [https://en.wikipedia.org/wiki/Antibody constant and variable domains] within the Fab region as antigen recognition occurs at the variable domain while the constant domain connects it to the rest of the IgM complex. Because the Fab region of IgM is poorly resolved, a structural analysis of an HIV neutralizing antibody called VCR01 was performed to approximate where an antigen would bind to at the <scene name='95/952714/Variable_region/1'>variable region</scene>. <ref name="Zhou">PMID:20616231</ref> |
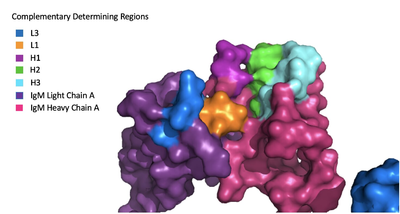
| - | The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes contact with the antibody on the Fab domain. Figure 2 depicts this as a surface representation | + | The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes contact with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. |
| - | Due to the poor resolution of the Fab region, specific side chain interactions between the heavy and light chains have not been determined. It is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. The <scene name='95/952713/Heavy-light_chain_interface/2'>heavy-light chain interface</scene> shows how the four heavy and light chain β-sandwiches fit together. The Fab region heavy chains attach to the Fc region heavy chains, before continuing down into the intracellular domain to interact with the Igα/Igβ subunits. The light chains however are only connected to the heavy chains within the Fab region, thus have no contact with the | + | Due to the poor resolution of the Fab region, specific side chain interactions between the heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chains have not been determined. It is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. The <scene name='95/952713/Heavy-light_chain_interface/2'>heavy-light chain interface</scene> shows how the four heavy and light chain β-sandwiches fit together. The Fab region heavy chains attach to the Fc region heavy chains, before continuing down into the intracellular domain to interact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> subunits. The light chains (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) however are only connected to the heavy chains ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) within the Fab region, thus have no contact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> heterodimer. |
[[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''' On one arm of the IgM antibody, the antigen makes contact with light chain A at the L1 and L3 complementary-determining regions. Furthermore, it makes contact with heavy chain A at the H1, H2, and H3 complementary-determining regions. The location of the complementary-determining regions were approximated using the structure of the VCR01 variable region and were visualized using Pymol.]] | [[Image:Igm_surface.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''' On one arm of the IgM antibody, the antigen makes contact with light chain A at the L1 and L3 complementary-determining regions. Furthermore, it makes contact with heavy chain A at the H1, H2, and H3 complementary-determining regions. The location of the complementary-determining regions were approximated using the structure of the VCR01 variable region and were visualized using Pymol.]] | ||
| Line 49: | Line 50: | ||
=='''Signal Transduction'''== | =='''Signal Transduction'''== | ||
| - | The diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown but it is speculated to operate under what is known as the conserved assembly mechanism | + | The diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown but it is speculated to operate under what is known as the conserved assembly mechanism. <ref name="Ma">PMID:35981028</ref> This means that upon antigen binding, BCRs on the surface of the cell begin to cluster to cause the phosphorylation of the immunoreceptor tyrosine-based activation motifs located in <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. In its “off” state, the constant region 4 of <b><span class="text-blue">heavy chain B</span></b> overlaps the extracellular components of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. As the antigen binds, it induces a conformational change to release the overlap and allow for clustering about the BCR. Now, in its “on” state the phosphorylation of the [https://en.wikipedia.org/wiki/Immunoreceptor_tyrosine-based_activation_motif ITAM region] (observed in figure 4 as conserved phosphorylated tyrosine residues) within the intracellular tails of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> drives downstream kinase activity to continue to process of [https://en.wikipedia.org/wiki/Tyrosine-protein_kinase_SYK signal cascading]. |
[[Image:Signal_diagram_2.png|400 px|left|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''' At the end of the intracellular Igα and Igβ helices are their cytoplasmic tails, and on each tail are tyrosine residues that are phosphorylated by one of two tyrosine kinase enzymes: Splenic-tyrosine kinase and Src family kinase. While the specific tyrosine residues are unknown in the mechanism, it is understood that their phosphorylation activates the B cell by triggering downstream intracellular signaling.]] | [[Image:Signal_diagram_2.png|400 px|left|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''' At the end of the intracellular Igα and Igβ helices are their cytoplasmic tails, and on each tail are tyrosine residues that are phosphorylated by one of two tyrosine kinase enzymes: Splenic-tyrosine kinase and Src family kinase. While the specific tyrosine residues are unknown in the mechanism, it is understood that their phosphorylation activates the B cell by triggering downstream intracellular signaling.]] | ||
{{Clear}} | {{Clear}} | ||
| - | |||
| - | =='''Medical Relevance'''== | ||
</StructureSection> | </StructureSection> | ||
| - | ==References== | + | =='''References'''== |
<references/> | <references/> | ||
| - | ==Student Contributors== | + | =='''Student Contributors'''== |
| - | + | DeTonyeá Dickson, | |
Allison Goss, | Allison Goss, | ||
Jackson Payton | Jackson Payton | ||
Revision as of 18:33, 7 April 2023
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
- ↑ Sathe A, Cusick JK. Biochemistry, Immunoglobulin M. 2022 Dec 19. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan–. PMID: 32310455. https://pubmed.ncbi.nlm.nih.gov/32310455/
- ↑ 2.0 2.1 Su Q, Chen M, Shi Y, Zhang X, Huang G, Huang B, Liu D, Liu Z, Shi Y. Cryo-EM structure of the human IgM B cell receptor. Science. 2022 Aug 19;377(6608):875-880. doi: 10.1126/science.abo3923. Epub 2022, Aug 18. PMID:35981043 doi:http://dx.doi.org/10.1126/science.abo3923
- ↑ 3.0 3.1 3.2 3.3 Ma X, Zhu Y, Dong, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. doi: 10.1126/science.abo3828. Epub 2022, Aug 18. PMID:35981028 doi:http://dx.doi.org/10.1126/science.abo3828
- ↑ 4.0 4.1 4.2 Tolar P, Pierce SK. Unveiling the B cell receptor structure. Science. 2022 Aug 19;377(6608):819-820. doi: 10.1126/science.add8065. Epub 2022 Aug 18.[http://dx.doi.org/10.1126/science.add8065 DOI:10.1126/science.add8065
- ↑ 5.0 5.1 5.2 Dylke J, Lopes J, Dang-Lawson M, Machtaler S, Matsuuchi L. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR). Immunol Lett. 2007 Sep 15;112(1):47-57. doi: 10.1016/j.imlet.2007.06.005. Epub 2007 Jul 23. [http://dx.doi.org/10.1016/j.imlet.2007.06.005 DOI:10.1016/j.imlet.2007.06.005
- ↑ Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010 Aug 13;329(5993):811-7. Epub 2010 Jul 8. PMID:20616231 doi:10.1126/science.1192819
Student Contributors
DeTonyeá Dickson, Allison Goss, Jackson Payton