We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1783
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Structural Overview == | == Structural Overview == | ||
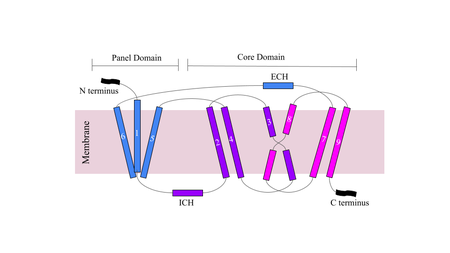
| - | NTCP has 9 transmembrane alpha helices (TM) that form the protein, with an extracellular N-terminus and intracellular C-terminus. NTCP has two domains within the protein, <scene name='95/952711/Panel_domain/1'>a panel domain</scene>, made up of TM1, TM5, and TM6, and <scene name='95/952711/Core_domain/3'>a core domain</scene>, made up of TM2-4 and TM7-9. An interesting feature of NTCP is the cross of TM3 and TM8 that form an <scene name='95/952711/X_motif/1'>X motif</scene> within the protein that is used in the conformational change of NTCP. The two domains are essential to the conformation change of NTCP to bind bile salts <Ref name="Xiangbing"> Xiangbing Qi, Wenhui Li. (2022). Unlocking the secrets to human NTCP structure. The Innovation, Vol. 3, Issue 5. 100294, ISSN 2666-6758, [https://doi.org/10.1016/j.xinn.2022.100294 DOI: 10.1016/j.xinn.2022.100294]. </Ref> | + | NTCP has 9 transmembrane alpha helices (TM) that form the protein, with an extracellular N-terminus and intracellular C-terminus. NTCP has two domains within the protein, <scene name='95/952711/Panel_domain/1'>a panel domain</scene>, made up of TM1, TM5, and TM6, and <scene name='95/952711/Core_domain/3'>a core domain</scene>, made up of TM2-4 and TM7-9. An interesting feature of NTCP is the cross of TM3 and TM8 that form an <scene name='95/952711/X_motif/1'>X motif</scene> within the protein that is used in the conformational change of NTCP. The two domains are essential to the conformation change of NTCP to bind bile salts <Ref name="Xiangbing"> Xiangbing Qi, Wenhui Li. (2022). Unlocking the secrets to human NTCP structure. The Innovation, Vol. 3, Issue 5. 100294, ISSN 2666-6758, [https://doi.org/10.1016/j.xinn.2022.100294 DOI: 10.1016/j.xinn.2022.100294]. </Ref> |
| - | There are two significant patches in the NTCP structure that facilitate ligand binding. Residues 84-87 of NTCP are <scene name='95/952711/Binding_patch_1/1'>Patch 1</scene>, which are located on the TM2-TM3 loop in the core domain. This patch is also considered the extracellular region of the binding tunnel within NTCP. Residues 157-165 of NTCP are associated with <scene name='95/952711/Binding_site_2_with_surface/1'>Patch 2</scene>. They are located on the N-terminal half of the TM5 in the panel domain (residue sequence: KGIVISLVL). Patch 2 is also located in the extracellular region of the binding tunnel. These residues' importance was determined through mutations of these residues and examined through pull-down assays <ref name="Asami"/> | + | There are two significant patches in the NTCP structure that facilitate ligand binding. Residues 84-87 of NTCP are <scene name='95/952711/Binding_patch_1/1'>Patch 1</scene>, which are located on the TM2-TM3 loop in the core domain. This patch is also considered the extracellular region of the binding tunnel within NTCP. Residues 157-165 of NTCP are associated with <scene name='95/952711/Binding_site_2_with_surface/1'>Patch 2</scene>. They are located on the N-terminal half of the TM5 in the panel domain (residue sequence: KGIVISLVL). Patch 2 is also located in the extracellular region of the binding tunnel. These residues' importance was determined through mutations of these residues and examined through pull-down assays. <ref name="Asami"/> The pre-S1 domain of HBV/HDV binds to the patches on NTCP in order to transport the virus from the exterior of NTCP to the interior binding tunnel to infect human liver cells.<ref name="Goutam"/> |
[[Image:Figuredomain.png|450 px|right|thumb|'''Figure 3.''' Cartoon of NTCP topology.]] | [[Image:Figuredomain.png|450 px|right|thumb|'''Figure 3.''' Cartoon of NTCP topology.]] | ||
| Line 27: | Line 27: | ||
=== Conformation Change === | === Conformation Change === | ||
| - | The conformational change of NTCP's core domain helices are essential to bile salt binding and uptake. Helices 3 and 8 <scene name='95/952711/X_motif/1'>of the X motif</scene> are the main structural components of the conformational change, as the X motif has highly conserved polar residue motifs that reside near the bile salt transport sites.. The conformational change is energized by the movement of Na+ down its concentration gradient. Before bile salt can bind, the pore in which salt binds must be <scene name='95/952711/Open_pore_ntcp_non_transparent/1'>open</scene>. Conserved glycine and proline residues act as hinges in the connecting short loops, intracellular α-helices, and extracellular α-helices of NTCP to facilitate the movement of the core and panel domain to allow for a conformational change. The <scene name='95/952711/Open_pore_ntcp/1'>open</scene> pore is flipped toward the outer membrane to allow for bile salt binding by exposing the Na+ binding sites and the X motif within NTCP. Once <scene name='95/952711/Open_pore_with_bile_salts/1'>bound</scene>, the pore is <scene name='95/952711/Closed_pore_ntcp/1'>closed</scene>, and bile salt is able to be released into the cell, past the inner membrane. | + | The conformational change of NTCP's core domain helices are essential to bile salt binding and uptake. Helices 3 and 8 <scene name='95/952711/X_motif/1'>of the X motif</scene> are the main structural components of the conformational change, as the X motif has highly conserved polar residue motifs that reside near the bile salt transport sites.. The conformational change is energized by the movement of Na+ down its concentration gradient. Before bile salt can bind, the pore in which salt binds must be <scene name='95/952711/Open_pore_ntcp_non_transparent/1'>open</scene>. Conserved glycine and proline residues act as hinges in the connecting short loops, intracellular α-helices, and extracellular α-helices of NTCP to facilitate the movement of the core and panel domain to allow for a conformational change. The <scene name='95/952711/Open_pore_ntcp/1'>open</scene> pore is flipped toward the outer membrane to allow for bile salt binding by exposing the Na+ binding sites and the X motif within NTCP. Once <scene name='95/952711/Open_pore_with_bile_salts/1'>bound</scene>, the pore is <scene name='95/952711/Closed_pore_ntcp/1'>closed</scene>, and bile salt is able to be released into the cell, past the inner membrane. <ref name="Goutam"/> |
=== Mechanism === | === Mechanism === | ||
| Line 37: | Line 37: | ||
== Significance == | == Significance == | ||
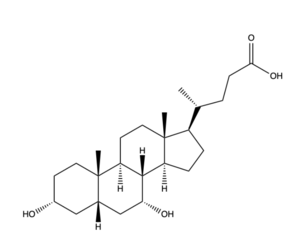
| - | NTCP is a key player in the absorption and digestion of fats and fat-soluble vitamins in the body, as well as the synthesis of bile within the liver. The uptake of bile salts, transcriptional and post-transcriptional, are signaling molecules for the liver and intestine. Bile salt, the transporting molecule of NTCP, aids in the absorption of lipophilic nutrients and vitamins in the small intestine. Additionally, bile salt plays a role in the endocrine system, excretion of toxins, and cholesterol maintenance. <ref name="Goutam"/> | + | NTCP is a key player in the absorption and digestion of fats and fat-soluble vitamins in the body, as well as the synthesis of bile within the liver. The uptake of bile salts, transcriptional and post-transcriptional, are signaling molecules for the liver and intestine. Bile salt, the transporting molecule of NTCP, aids in the absorption of lipophilic nutrients and vitamins in the small intestine. Additionally, bile salt plays a role in the endocrine system, excretion of toxins, and cholesterol maintenance. <ref name="Goutam"/> Thus, the significance of NTCP lies with the importance of bile salts; NTCP is necessary in maintaining bile salt levels and thus necessary in maintaining the aforementioned biological processes. |
=== Bile Salt Uptake === | === Bile Salt Uptake === | ||
Revision as of 20:40, 10 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
Sodium Taurocholate Co-Transporting Peptide
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 1.4 Goutam, K., Ielasi, F.S., Pardon, E. et al. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606, 1015–1020 (2022). DOI: 10.1038/s41586-022-04723-z.
- ↑ Maldonado-Valderrama, J., Wilde, P., Macierzanka, A., & Mackie, A. (2011). The role of bile salts in digestion. Advances in colloid and interface science, 165(1), 36–46. DOI: 10.1016/j.cis.2010.12.002.
- ↑ 3.0 3.1 Asami J, Kimura KT, Fujita-Fujiharu Y, Ishida H, Zhang Z, Nomura Y, Liu K, Uemura T, Sato Y, Ono M, Yamamoto M, Noda T, Shigematsu H, Drew D, Iwata S, Shimizu T, Nomura N, Ohto U. Structure of the bile acid transporter and HBV receptor NTCP. Nature. 2022 Jun; 606 (7916):1021-1026. DOI: 10.1038/s41586-022-04845-4.
- ↑ Xiangbing Qi, Wenhui Li. (2022). Unlocking the secrets to human NTCP structure. The Innovation, Vol. 3, Issue 5. 100294, ISSN 2666-6758, DOI: 10.1016/j.xinn.2022.100294.
- ↑ Liu, H., Irobalieva, R.N., Bang-Sørensen, R. et al. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res 32, 773–776 (2022). DOI: 10.1038/s41422-022-00680-4.
- ↑ Vlahcevic, Z., Buhac, I., et al. Bile Acid Metabolism in Patients with Cirrhosis. Gastroenterology vol. 60, 491-498 (1971). DOI: 10.1016/S0016-5085(71)80053-7.
![Figure 1. NTCP structure with both Na+ ions and bile salts bound with the NTCP molecule shown in light blue, the two Na+ ions shown in purple spheres, and the bile salts shown in sticks as dark blue. [PDB file 7ZYI].](/wiki/images/thumb/3/36/Fig_1.png/400px-Fig_1.png)