BASIL2023GV1ZBS
From Proteopedia
| Line 1: | Line 1: | ||
=='''''Inquiry of the Possible Function of Protein 1ZBS'''''== | =='''''Inquiry of the Possible Function of Protein 1ZBS'''''== | ||
| + | ==''' Abstract'''== | ||
| + | ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity. | ||
== '''Introduction''' == | == '''Introduction''' == | ||
<StructureSection load='1zbs' size='340' side='right' caption='1ZBS dimer as in its original state' scene=''> | <StructureSection load='1zbs' size='340' side='right' caption='1ZBS dimer as in its original state' scene=''> | ||
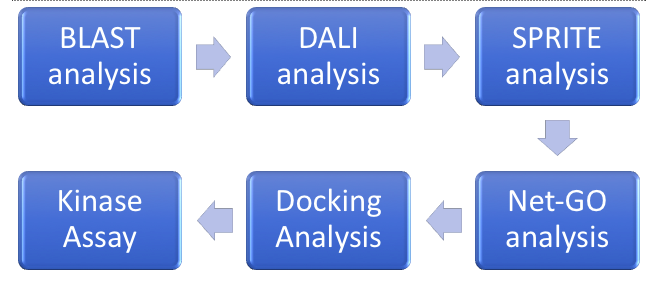
1ZBS is a protein with unknown function discovered through a genomics project of predictive folding. The goal of this project was to determine what the function of protein 1ZBS was using a combination of computational analysis and physical lab work. | 1ZBS is a protein with unknown function discovered through a genomics project of predictive folding. The goal of this project was to determine what the function of protein 1ZBS was using a combination of computational analysis and physical lab work. | ||
[[Image: FlowChart.png ]] | [[Image: FlowChart.png ]] | ||
| - | For this project, we started by doing our computational work, which includes BLAST, DALI, SPRITE, Net-GO, and the Docking analysis, while we started transforming our bacteria, and over-expressing and purifying our protein for the kinase assays. | + | For this project, we started by doing our computational work, which includes BLAST, DALI, SPRITE, Net-GO, and the Docking analysis, while we started transforming our bacteria, and over-expressing and purifying our protein for the kinase assays. From the computational analysis, it was believed that 1ZBS could be a NAGK protein. Further analysis into the kinase activity did not fully support this conclusion, as the specific activity was too low. |
== '''Sequence Alignments''' == | == '''Sequence Alignments''' == | ||
| Line 12: | Line 14: | ||
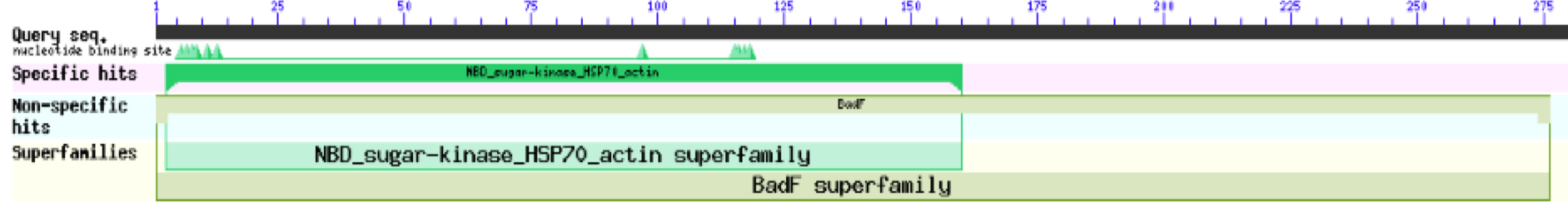
The first type of analysis that was done was using Protein Blast from NCBI. The analysis was first done as it only gave very broad information about the potential function of the protein, by matching domains across other proteins with known functions. | The first type of analysis that was done was using Protein Blast from NCBI. The analysis was first done as it only gave very broad information about the potential function of the protein, by matching domains across other proteins with known functions. | ||
[[Image:BLASTP.png]] | [[Image:BLASTP.png]] | ||
| + | These results for 1ZBS showed that it was a match to the superfamily BADF. BADF is the superfamily that NAGK proteins also belong in so this was one of the reasons that we believed that 1ZBS may potentially be a NAGK protein. | ||
== '''Structural Alignments''' == | == '''Structural Alignments''' == | ||
| - | + | === DALI === | |
| + | DALI is a global alignment software that is used to match proteins up to possible structural matches within the Protein Data Bank (PDB). | ||
| + | [[Image:DALI_alignment.png]] | ||
| + | This is the DALI alignment of 1ZBS and 2CH5, where 1ZBS is colored in green and 2CH5 is colored in yellow. The best results from the DALI alignment were mainly proteins with unknown functions, which makes it difficult to deduce a possible function off of. Because of these results, 2CH5, while only matching parts of the sequence was the best result for this analysis as it's function is known. The function of 2CH5 is a NAG kinase, which would further support the thought that 1ZBS could also be a NAG kinase. | ||
==''' Substrate Possibilities '''== | ==''' Substrate Possibilities '''== | ||
Revision as of 19:01, 11 April 2023
Contents |
Inquiry of the Possible Function of Protein 1ZBS
Abstract
ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity.
Introduction
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Danielle Selover, Carmen Almendarez Rodriguez, Bonnie Hall, Jaime Prilusky