We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1777
From Proteopedia
(Difference between revisions)
| Line 39: | Line 39: | ||
==Active Site of PP1C== | ==Active Site of PP1C== | ||
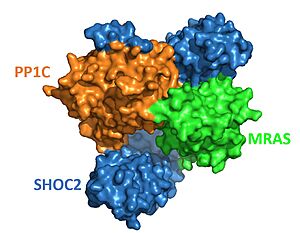
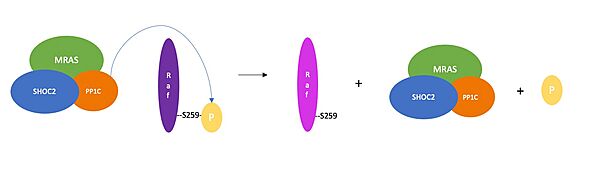
| - | Once all the domains are bound, [https://en.wikipedia.org/wiki/Nucleoside_triphosphate NTpS] binds to PP1C in the active site. NTpS refers to the phosphorylated serine (residue 259) located at the N-terminus of RAF. Once NTpS is bound, PP1C catalyzes the dephosphorylation of serine 259. NTpS is dephosphorylated to prevent the active dimeric RAF from inactivating and changing into its monomeric structure. Notably, NTpS can bind to the PP1C active site without PP1C being in complex with SHOC2 and MRAS. This occurs when RAS binds to RAF, which exposes NTpS on RAF. However, for this reaction, the catalytic activity of PP1C outside of the complex is less efficient. | + | Once all the domains are bound, [https://en.wikipedia.org/wiki/Nucleoside_triphosphate NTpS] binds to PP1C in the active site. NTpS refers to the phosphorylated serine (residue 259) located at the N-terminus of RAF. Once NTpS is bound, PP1C catalyzes the dephosphorylation of serine 259. NTpS is dephosphorylated to prevent the active dimeric RAF from inactivating and changing into its monomeric structure. Notably, NTpS can bind to the PP1C active site without PP1C being in complex with SHOC2 and MRAS. This occurs when RAS binds to RAF, which exposes NTpS on RAF. However, for this reaction, the catalytic activity of PP1C outside of the complex is less efficient. It's hypothesized this is because hydrophobic residues directly C-terminal to the phosphorylated serine bind to the hydrophobic patch of PP1C as well as the hydrophobic SHOC2 C-terminus<ref name="Liau">PMID:35768504</ref>. The conserved hydrophobic groove at the SHOC2 C-terminus and the hydrophobic region next to the PP1C active site neighbor each other in the structure of the complex. As a result, the hydrophobic residues downstream from NTpS bind to the hydrophobic path of PP1C next to the active site and to the hydrophobic groove in the SHOC2 C-terminus. '''<scene name='95/952705/Pp1c_active_site/4'>NTpS</scene> is surrounded by hydrophobic and acidic regions in the C-terminal direction. Similarly, hydrophobic regions are located on the surface of PP1C, whereas the active site is placed further into the structure of PP1C due to its charged nature. It is thought that these regions help NTpS bind to the active site by making interactions that will lead NTpS into the protein. There is still some uncertainty as to how the substrate selectivity works, but these regions could play an essential role in it.<ref name="Liau">PMID:35768504</ref>.''' |
| - | + | ||
| - | ''' | + | |
| - | NTpS can bind to the PP1C active site without PP1C also being bound to SHOC2 and MRAS, however, the catalytic activity is much slower and the reaction is less efficient. In order for NTpS to bind PP1C in the absence of the other two proteins, NTpS would need to be exposed from its binding site in the inactive RAF complex. RAS has to bind to RAF to expose the NTpS, allowing PP1C and NTpS to bind. | + | '''NTpS can bind to the PP1C active site without PP1C also being bound to SHOC2 and MRAS, however, the catalytic activity is much slower and the reaction is less efficient. In order for NTpS to bind PP1C in the absence of the other two proteins, NTpS would need to be exposed from its binding site in the inactive RAF complex. RAS has to bind to RAF to expose the NTpS, allowing PP1C and NTpS to bind.''' |
<scene name='95/952705/Pp1c_active_site/5'>TextToBeDisplayed</scene> '''let me know thought on the style of image''' | <scene name='95/952705/Pp1c_active_site/5'>TextToBeDisplayed</scene> '''let me know thought on the style of image''' | ||
Revision as of 12:36, 17 April 2023
| This Sandbox is Reserved from February 27 through August 31, 2023 for use in the course CH462 Biochemistry II taught by R. Jeremy Johnson at the Butler University, Indianapolis, USA. This reservation includes Sandbox Reserved 1765 through Sandbox Reserved 1795. |
To get started:
More help: Help:Editing |
| |||||||||||
References
- ↑ 1.0 1.1 Bernal Astrain G, Nikolova M, Smith MJ. Functional diversity in the RAS subfamily of small GTPases. Biochem Soc Trans. 2022 Apr 29;50(2):921-933. doi: 10.1042/BST20211166. DOI:10.1042/BST20211166
- ↑ 2.0 2.1 Molina JR, Adjei AA. The Ras/Raf/MAPK pathway. J Thorac Oncol. 2006 Jan;1(1):7-9. DOI:10.1016/S1556-0864(15)31506-9.
- ↑ Li, L., Zhao, G. D., Shi, Z. et. al.The Ras/Raf/MEK/ERK signaling pathway (Figure 1) and its role in the occurrence and development of HCC. Oncology letters, 12(5), 3045–3050. DOI:10.3892/ol.2016.5110.
- ↑ 4.0 4.1 4.2 Hauseman, Z.J., Fodor, M., Dhembi, A. et al. Structure of the MRAS–SHOC2–PP1C phosphatase complex. Nature 609, 416–423 (2022). doi: 10.1038/s41586-022-05086-1. DOI:10.1038/s41586-022-05086-1.
- ↑ 5.0 5.1 Kwon, J. J., & Hahn, W. C. A Leucine-Rich Repeat Protein Provides a SHOC2 the RAS Circuit: a Structure-Function Perspective. Molecular and cellular biology, 41(4), e00627-20 (2021). doi:10.1128/MCB.00627-20. DOI: 10.1128/MCB.00627-20.
- ↑ Zhou, Y., Prakash, P., Liang, H., et al. Lipid-Sorting Specificity Encoded in K-Ras Membrane Anchor Regulates Signal Output. Cell, 168(1-2), 239–251.e16 doi: 10.1016/j.cell.2016.11.059. DOI: 10.1016/j.cell.2016.11.059.
- ↑ Young, L., Rodriguez-Viciana, P. MRAS: A Close but Understudied Member of the RAS Family. Cold Spring Harbor Perspectives in Medicine (2018). doi: 10.1101/cshperspect.a033621. DOI: 0.1101/cshperspect.a033621.
- ↑ Daniel A. Bonsor, Patrick Alexander, Kelly Snead, Nicole Hartig, Matthew Drew, Simon Messing, Lorenzo I. Finci, Dwight V. Nissley, Frank McCormick, Dominic Esposito, Pablo Rodrigiguez-Viciana, Andrew G. Stephen, Dhirendra K. Simanshu. Structure of the SHOC2–MRAS–PP1C complex provides insights into RAF activation and Noonan syndrome. bioRxiv. 2022.05.10.491335. doi: 10.1101/2022.05.10.491335. DOI:10.1101/2022.05.10.491335.
- ↑ 9.0 9.1 9.2 9.3 9.4 Kwon, J.J., Hajian, B., Bian, Y. et al. Structure–function analysis of the SHOC2–MRAS–PP1C holophosphatase complex. Nature 609, 408–415 (2022).doi: 10.1038/s41586-022-04928-2. DOI:10.1038/s41586-022-04928-2
- ↑ 10.0 10.1 10.2 Kwon, J., Jajian, B., Bian, Y. et al. Comprehensive structure-function evaluation of the SHOC2 holophosphatase reveals disease mechanisms and therapeutic opportunities. In: Proceedings of the American Association for Cancer Research Annual Meeting 2022. DOI: 10.1158/1538-7445.AM2022-LB029.
- ↑ 11.0 11.1 Liau NPD, Johnson MC, Izadi S, Gerosa L, Hammel M, Bruning JM, Wendorff TJ, Phung W, Hymowitz SG, Sudhamsu J. Structural basis for SHOC2 modulation of RAS signalling. Nature. 2022 Jun 29. pii: 10.1038/s41586-022-04838-3. doi:, 10.1038/s41586-022-04838-3. PMID:35768504 doi:http://dx.doi.org/10.1038/s41586-022-04838-3
- ↑ 12.0 12.1 Lavoie, H., Therrien, M. Structural keys unlock RAS–MAPK cellular signaling pathway. Nature 609, 248-249 (2022). doi: 10.1038/d41586-022-02189-7. DOI:10.1038/d41586-022-02189-7.
- ↑ 13.0 13.1 van der Burgt, I. Noonan syndrome. Orphanet J Rare Dis 2, 4 (2007). doi: 10.1186/1750-1172-2-4 DOI: 10.1186/1750-1172-2-4.