BASIL2023GV1ZBS
From Proteopedia
| Line 26: | Line 26: | ||
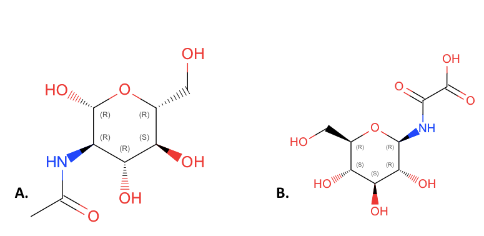
From there we used PyRx to do molecular docking into the 1ZBS protein. This was completed using various different substrates in combination with ATP, which is needed for the kinase to be able to function. The control used was Imidazole which had a binding affinity of -2.9 kcal/mol. By completing this first, we had a better idea of what our binding affinity should be. Then we used <scene name='95/957643/Nag/1'>NAG</scene> to better determine if it was our substrate, as well as other similar structures found using the PDB. NAG had a binding affinity of -5.54kcal/mol. This was a better result than our control, and this helped further the belief that 1ZBS could be a NAG kinase. | From there we used PyRx to do molecular docking into the 1ZBS protein. This was completed using various different substrates in combination with ATP, which is needed for the kinase to be able to function. The control used was Imidazole which had a binding affinity of -2.9 kcal/mol. By completing this first, we had a better idea of what our binding affinity should be. Then we used <scene name='95/957643/Nag/1'>NAG</scene> to better determine if it was our substrate, as well as other similar structures found using the PDB. NAG had a binding affinity of -5.54kcal/mol. This was a better result than our control, and this helped further the belief that 1ZBS could be a NAG kinase. | ||
| - | The docking results did also give us a substrate with much more desirable binding affinity, which was N-(carboxycarbonyl)-glucosylamine, otherwise referred to as 4GP. This substrate had a binding affinity of -6.08 kcal/mol. | + | The docking results did also give us a substrate with much more desirable binding affinity, which was N-(carboxycarbonyl)-glucosylamine, otherwise referred to as <scene name='95/957643/4gp/1'>4GP</scene>. This substrate had a binding affinity of -6.08 kcal/mol. |
==''' Substrate Possibilities '''== | ==''' Substrate Possibilities '''== | ||
Revision as of 17:39, 19 April 2023
Contents |
Inquiry of the Possible Function of Protein 1ZBS
Abstract
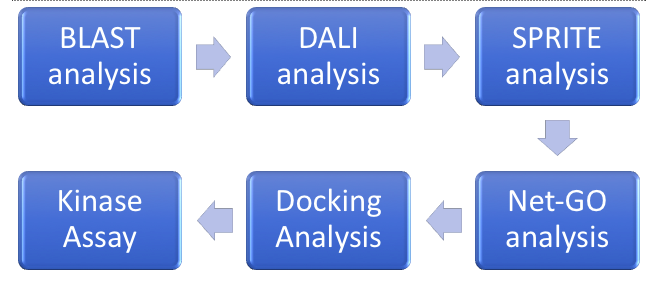
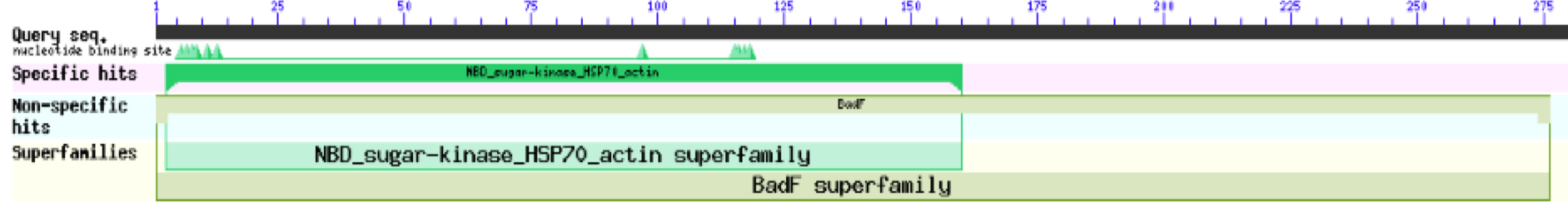
ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity.
Introduction
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Danielle Selover, Carmen Almendarez Rodriguez, Bonnie Hall, Jaime Prilusky