We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1789
From Proteopedia
(Difference between revisions)
| Line 24: | Line 24: | ||
=== PP1C === | === PP1C === | ||
| + | <scene name='95/952717/Pp1c/1'>PP1C</scene> is a phosphatase. After forming a ternary complex, the hydrophobic active site on PP1C interacts with Raf and dephosphorylate Ser 259. PP1C's active site is adjacent to a hydrophobic binding pocket that binds to the C-terminal phosphoserine, located on the N-terminus of RAF, the target for dephosphorylation. PP1C can act as a phosphatase in the absence of SHOC2 but PP1C lacks intrinsic substrate selectively. The SMP complex formation endows PP1C with specificity for RAF <ref name="Hauseman" />. | ||
| + | |||
| + | The mechanism that PP1C uses to catalyze the dephosphorylation is mainly through donating a hydrogen atom to a phosphate group on the C-terminal of a [https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/phosphoserine. phosphoserine] on Raf <ref name="Kubicek"/>. This makes the phosphate group a good leaving group and it breaks off <ref name="Kubicek"/>. This catalysis is done by the serine-threonine alpha catalytic site on PP1C <ref name="Kubicek"/>. In this catalytic site, there are two Manganese ions and one calcium ion <ref name="Kubicek"/>. These metal ions are necessary in stablizing this catalytic site and there are a lot of polar negative residues in this region <ref name="Kubicek"/> | ||
| + | |||
| + | |||
<scene name='95/952717/Pp1c/1'>PP1C</scene> | <scene name='95/952717/Pp1c/1'>PP1C</scene> | ||
=== MRAS === | === MRAS === | ||
| + | |||
| + | <scene name='95/952717/Mras/2'>MRAS</scene> is a monomeric GTPase. MRAS is membrane-bound due to post-translational lipidation which allows the protein to interact with the inner membrane leaflet. <ref name="Seabra">PMID:9607139</ref> MRAS localizes the SMP complex near RAF and other components of downstream signaling. The region of MRAS not directly bound to the membrane binds SHOC2 and PP1C to orient the complex such that PP1C’s active site faces the serine that will get dephosphorylated on RAF. MRAS also controls SMP complex formation in connection with extracellular signaling based on its dualistic switching between its inactive and active state. In its inactive state, MRAS is bound to GDP. When signaled by growth factors, the GDP is exchanged for GTP when a ligand binds to the RTK <ref name="Hauseman" />. The now <scene name='95/952718/Zoom_in_gtp/1'>GTP bound MRAS</scene> undergoes a conformational change of the <scene name='95/952716/Ras-switch-zoomed/1'>switch I and switch II regions</scene>. These regions are the major binding sites with SHOC2. This conformational change activates MRAS allowing it to bind with the SHOC2-PP1C complex. In its inactive GDP-bound state, MRAS is sterically occluded from binding SHOC2. For example, R83 of GDP-bound MRAS directly clashes with SHOC2 as shown in figure 2. In comparison to other RAS proteins such as H/K/NRAS, MRAS has a greater affinity for the SHOC2-PP1C complex<ref name="Kubicek">Kubicek M, Pacher M, Abraham D, Podar K, Eulitz M, Baccarini M. Dephosphorylation of Ser-259 regulates Raf-1 membrane association. J Biol Chem. 2002 Mar 8;277(10):7913-9. [http://10.1074/jbc.M108733200 doi: 10.1074/jbc.M108733200.]</ref>. This indicates that the specific structure of MRAS is necessary for SMP function. While MRAS engages the SHOC2-PP1C complex to bring the complex to the membrane, an additional membrane-bound RAS binds RAF nearby. This binding is also stimulated by ligand binding to the RTK. This indicates that for full RAF activation and continuous signaling of Raf, two separate active RAS proteins are needed. Having two MRASs also help with the co-localization of PP1C to the NTpS region on RAF. To inactivate Raf signaling, MRAS uses its intrinsic GTPase to remove the activating gamma-phosphate on GTP. In the GDP-bound state, switch I and II move to the position shown in green in Figure 2. This inactivates SHOC2 binding due to steric clashing which causes the SMP structure to dissociate. | ||
<scene name='95/952717/Mras/2'>MRAS</scene> | <scene name='95/952717/Mras/2'>MRAS</scene> | ||
<scene name='95/952716/Gtp/1'>GTP-MRAS(full-image)</scene> | <scene name='95/952716/Gtp/1'>GTP-MRAS(full-image)</scene> | ||
| Line 31: | Line 38: | ||
<scene name='95/952716/Ras-swtich/2'>Switch I-II (full-image)</scene> | <scene name='95/952716/Ras-swtich/2'>Switch I-II (full-image)</scene> | ||
<scene name='95/952716/Ras-switch-zoomed/1'>Switch I-II (zoomed-in)</scene> | <scene name='95/952716/Ras-switch-zoomed/1'>Switch I-II (zoomed-in)</scene> | ||
| + | |||
=== Key Ligand Interactions === | === Key Ligand Interactions === | ||
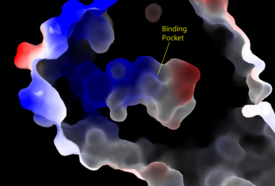
[[Image:Amphbindingfinal.png|275 px|right|thumb|'''Figure 3''': Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] | [[Image:Amphbindingfinal.png|275 px|right|thumb|'''Figure 3''': Electrostatic illustration of the amphipathic binding pocket of the LPA<sub>1</sub> receptor. This binding pocket was revealed by cutting away the exterior or the protein. This binding pocket, located in the interior of the protein, has both polar and nonpolar regions. The blue and red coloration highlight the positively and negatively charged regions, respectively, and the white color shows the nonpolar region of the binding pocket.]] | ||
Revision as of 19:12, 19 April 2023
This page, as it appeared on June 14, 2016, was featured in this article in the journal Biochemistry and Molecular Biology Education.
SHOC2-PP1C-MRAS
| |||||||||||
Student Contributors
Madeline Gilbert Inaya Patel Rushda Hussein