We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Sandbox Reserved 1785
From Proteopedia
(Difference between revisions)
| Line 48: | Line 48: | ||
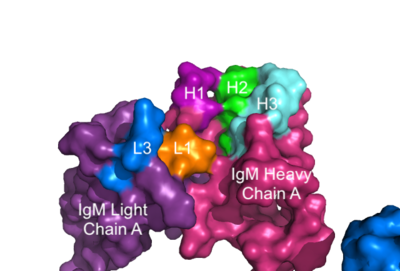
The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes <scene name='95/952713/Antigen_antibody_contact/4'>contact</scene> with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. | The IgM-BCR contains areas referred to as [https://en.wikipedia.org/wiki/Complementarity-determining_region complementary-determining regions](CDRs), which are where the antigen makes <scene name='95/952713/Antigen_antibody_contact/4'>contact</scene> with the antibody on the Fab domain. Figure 2 depicts this as a surface representation given that the specific residues within the antigen-binding motif are unknown. | ||
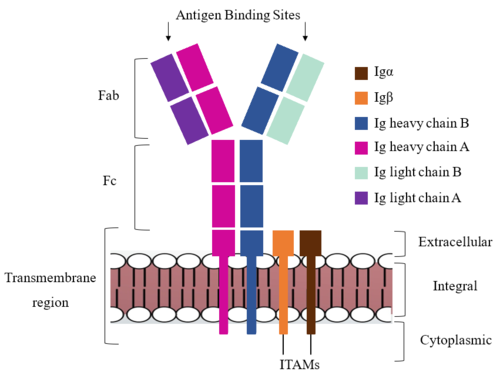
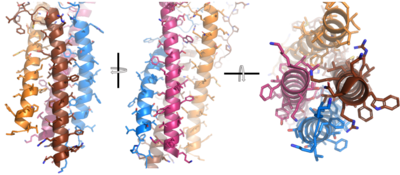
| - | Due to the poor resolution of the Fab region, specific side chain interactions between the heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chains have not been determined. Based on the general structure of an [https://en.wikipedia.org/wiki/Antibody antibody] it is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. The poor resolution does not allow them to be visualized within the structure, but upon understanding the significance of cellular signals being transmitted down to the Fc region, the presence of disulfide bonds would allow the protein to maintain its stability during conformational changes that occur as a result of signal modulation. The <scene name='95/952713/Heavy-light_chain_interface/2'>heavy-light chain interface</scene> shows how the four heavy and light chain β-sandwiches fit together. The Fab region heavy chains attach to the Fc region heavy chains, before continuing down into the intracellular domain to interact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> subunits. The light chains (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) however are only connected to the heavy chains ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) within the Fab region, thus have no contact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> heterodimer. | + | Due to the poor resolution of the Fab region, specific side chain interactions between the heavy ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) and light (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) chains have not been determined. Based on the general structure of an [https://en.wikipedia.org/wiki/Antibody antibody] it is estimated that each β-sandwich contains one disulfide bridge with additional hydrogen bonds. <ref name="Su">PMID:35981043</ref> The poor resolution does not allow them to be visualized within the structure, but upon understanding the significance of cellular signals being transmitted down to the Fc region, the presence of disulfide bonds would allow the protein to maintain its stability during conformational changes that occur as a result of signal modulation. The <scene name='95/952713/Heavy-light_chain_interface/2'>heavy-light chain interface</scene> shows how the four heavy and light chain β-sandwiches fit together. The Fab region heavy chains attach to the Fc region heavy chains, before continuing down into the intracellular domain to interact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> subunits. The light chains (<b><span class="text-purple">A</span></b>/<b><span class="text-cyan">B</span></b>) however are only connected to the heavy chains ('''{{Font color|violet|A}}'''/<b><span class="text-blue">B</span></b>) within the Fab region, thus have no contact with the <b><span class="text-brown">Igα</span></b>/<b><span class="text-orange">Igβ</span></b> heterodimer. |
[[Image:IgM_Surface2.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''' On one arm of the IgM antibody, the antigen makes contact with light chain A at the L1 and L3 complementary-determining regions. The antigen also makes contact with heavy chain A at the H1, H2, and H3 complementary-determining regions. The location of the complementary-determining regions were approximated using the structure of the VCR01 variable region and were visualized using Pymol.]] | [[Image:IgM_Surface2.png|400 px|left|thumb|'''Figure 3. Surface Representation of IgM Antibody Binding Pocket.''' On one arm of the IgM antibody, the antigen makes contact with light chain A at the L1 and L3 complementary-determining regions. The antigen also makes contact with heavy chain A at the H1, H2, and H3 complementary-determining regions. The location of the complementary-determining regions were approximated using the structure of the VCR01 variable region and were visualized using Pymol.]] | ||
| Line 57: | Line 57: | ||
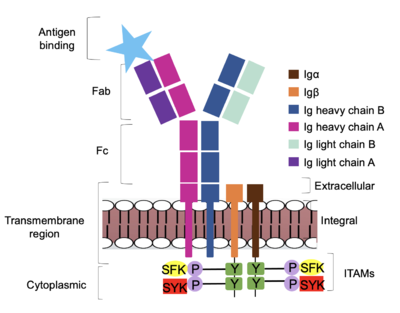
[[Image:Signal_diagram_2.png|400 px|right|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''' The tyrosine residues on the intracellular end of Igα and Igβ helices are phosphorylated by potential tyrosine kinases: Splenic-tyrosine kinase and Src family kinase. Although the exact mechanism is unknown, the phosphorylation of Igα and Igβ activates the B cell and triggers intracellular downstream signaling.]] | [[Image:Signal_diagram_2.png|400 px|right|thumb|'''Figure 4. IgM Antibody Signal Transduction following Antigen Binding.''' The tyrosine residues on the intracellular end of Igα and Igβ helices are phosphorylated by potential tyrosine kinases: Splenic-tyrosine kinase and Src family kinase. Although the exact mechanism is unknown, the phosphorylation of Igα and Igβ activates the B cell and triggers intracellular downstream signaling.]] | ||
| - | <div style="text-align: justify">Based on the structure of IgM, the diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown due to the lack of specificity on which tyrosine residues in the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> domains are phosphorylated, but it is speculated to operate under what is known as the conserved assembly mechanism. <ref name="Ma">PMID:35981028</ref> Following an antigen binding, BCRs on the surface of the cell begin to cluster to cause the phosphorylation of the ITAMs located in <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. In its “off” state, the constant region 4 of <b><span class="text-blue">heavy chain B</span></b> <scene name='95/952713/Signal_clustering/2'>overlaps</scene> the extracellular components of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. The antigen binding induces a conformational change to release the overlap and allow for clustering about the BCR. Now, in its “on” state, the phosphorylation of the [https://en.wikipedia.org/wiki/Immunoreceptor_tyrosine-based_activation_motif ITAM region] within the intracellular tails of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> drives downstream kinase activity. As observed in Figure 4, the activated antibody can now continue to process of [https://en.wikipedia.org/wiki/Tyrosine-protein_kinase_SYK intracellular signal cascading]. | + | <div style="text-align: justify">Based on the structure of IgM, the diagram in Figure 4 depicts the initial process of B cell activation by the antigen binding to the antibody at the Fab region. The underlying mechanism for signal transduction is unknown due to the lack of specificity on which tyrosine residues in the <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> domains are phosphorylated, but it is speculated to operate under what is known as the conserved assembly mechanism. <ref name="Ma">PMID:35981028</ref> Following an antigen binding, BCRs on the surface of the cell begin to cluster to cause the phosphorylation of the ITAMs located in <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. In its “off” state, the constant region 4 of <b><span class="text-blue">heavy chain B</span></b> <scene name='95/952713/Signal_clustering/2'>overlaps</scene> the extracellular components of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b>. The antigen binding induces a conformational change to release the overlap and allow for clustering about the BCR. Now, in its “on” state, the phosphorylation of the [https://en.wikipedia.org/wiki/Immunoreceptor_tyrosine-based_activation_motif ITAM region] within the intracellular tails of <b><span class="text-brown">Igα</span></b> and <b><span class="text-orange">Igβ</span></b> drives downstream kinase activity. As observed in Figure 4, the activated antibody can now continue to process of [https://en.wikipedia.org/wiki/Tyrosine-protein_kinase_SYK intracellular signal cascading]. <ref name="Ma">PMID:35981028</ref> |
</StructureSection> | </StructureSection> | ||
Current revision
Human B-cell Antigen Receptor: IgM BCR
| |||||||||||
References
- ↑ Sathe A, Cusick JK. Biochemistry, Immunoglobulin M. 2022 Dec 19. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan–. PMID: 32310455. https://pubmed.ncbi.nlm.nih.gov/32310455/
- ↑ 2.0 2.1 2.2 Su Q, Chen M, Shi Y, Zhang X, Huang G, Huang B, Liu D, Liu Z, Shi Y. Cryo-EM structure of the human IgM B cell receptor. Science. 2022 Aug 19;377(6608):875-880. doi: 10.1126/science.abo3923. Epub 2022, Aug 18. PMID:35981043 doi:http://dx.doi.org/10.1126/science.abo3923
- ↑ 3.0 3.1 3.2 3.3 3.4 Ma X, Zhu Y, Dong, Chen Y, Wang S, Yang D, Ma Z, Zhang A, Zhang F, Guo C, Huang Z. Cryo-EM structures of two human B cell receptor isotypes. Science. 2022 Aug 19;377(6608):880-885. doi: 10.1126/science.abo3828. Epub 2022, Aug 18. PMID:35981028 doi:http://dx.doi.org/10.1126/science.abo3828
- ↑ 4.0 4.1 4.2 Tolar P, Pierce SK. Unveiling the B cell receptor structure. Science. 2022 Aug 19;377(6608):819-820. doi: 10.1126/science.add8065. Epub 2022 Aug 18.[http://dx.doi.org/10.1126/science.add8065 DOI:10.1126/science.add8065
- ↑ 5.0 5.1 5.2 Dylke J, Lopes J, Dang-Lawson M, Machtaler S, Matsuuchi L. Role of the extracellular and transmembrane domain of Ig-alpha/beta in assembly of the B cell antigen receptor (BCR). Immunol Lett. 2007 Sep 15;112(1):47-57. doi: 10.1016/j.imlet.2007.06.005. Epub 2007 Jul 23. [http://dx.doi.org/10.1016/j.imlet.2007.06.005 DOI:10.1016/j.imlet.2007.06.005
- ↑ Daniels R, Kurowski B, Johnson AE, Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003 Jan;11(1):79-90. PMID:12535523 doi:10.1016/s1097-2765(02)00821-3
- ↑ Bakshi T, Pham D, Kaur R, Sun B. Hidden Relationships between N-Glycosylation and Disulfide Bonds in Individual Proteins. Int J Mol Sci. 2022 Mar 29;23(7):3742. PMID:35409101 doi:10.3390/ijms23073742
- ↑ Mirazimi A, Svensson L. Carbohydrates facilitate correct disulfide bond formation and folding of rotavirus VP7. J Virol. 1998 May;72(5):3887-92. PMID:9557673 doi:10.1128/JVI.72.5.3887-3892.1998
- ↑ Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Do Kwon Y, Scheid JF, Shi W, Xu L, Yang Y, Zhu J, Nussenzweig MC, Sodroski J, Shapiro L, Nabel GJ, Mascola JR, Kwong PD. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010 Aug 13;329(5993):811-7. Epub 2010 Jul 8. PMID:20616231 doi:10.1126/science.1192819
Student Contributors
DeTonyeá Dickson, Allison Goss, Jackson Payton