BASIL2023GV1ZBS
From Proteopedia
| Line 28: | Line 28: | ||
=== NetGO === | === NetGO === | ||
| - | We used NetGo 2.0 to help visualize the potential function of 1ZBS. This software uses predictive modeling to determine potential functions of the protein in query, and produces a set of (GO) terms that best aligns with the protein's possible function. | + | We used NetGo 2.0 to help visualize the potential function of 1ZBS. This software uses predictive modeling to determine potential functions of the protein in query, and produces a set of gene ontology(GO) terms that best aligns with the protein's possible function. It starts with the most general information and builds to become more specific. These results came back in a list form. We then used AmiGo to visualize the connection between the terms. The final results landed us with a protein that has a purine ribonucleotide triphosphate binding domain, and could have some NAG kinase activity. |
==''' Docking Analysis'''== | ==''' Docking Analysis'''== | ||
| Line 49: | Line 49: | ||
== '''Conclusion''' == | == '''Conclusion''' == | ||
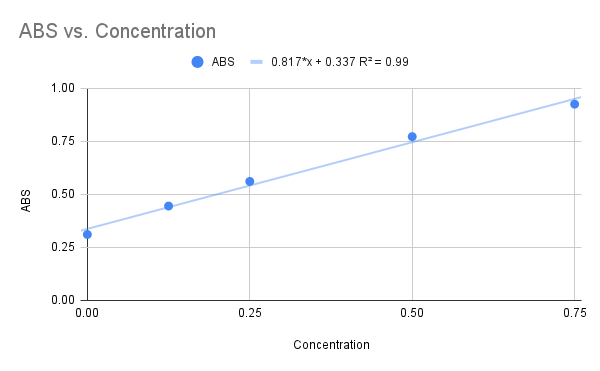
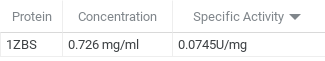
The goal of this experiment was to determine the function of 1zbs. We did this by creating a standard curve of BSA to compare our curve to our possible substrate NAG. This was done by a series of dilutions and Bradford assays to obtain a dilution factor by comparing it the absorbance levels of BSA. We concluded from our kinase assay results that 1ZBS is not a kinase because the specific activity was 0.074 U/mg. | The goal of this experiment was to determine the function of 1zbs. We did this by creating a standard curve of BSA to compare our curve to our possible substrate NAG. This was done by a series of dilutions and Bradford assays to obtain a dilution factor by comparing it the absorbance levels of BSA. We concluded from our kinase assay results that 1ZBS is not a kinase because the specific activity was 0.074 U/mg. | ||
| - | This is a sample scene created with SAT to <scene name="/12/3456/Sample/1">color</scene></scene> by Group, and another to make <scene name='95/957643/1zbs_view_2/1'> a transparent representation</scene> of the protein. You can make your own scenes on SAT starting from scratch or loading and editing one of these sample scenes. | ||
</StructureSection> | </StructureSection> | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 22:35, 22 April 2023
Contents |
Inquiry of the Possible Function of Protein 1ZBS
Abstract
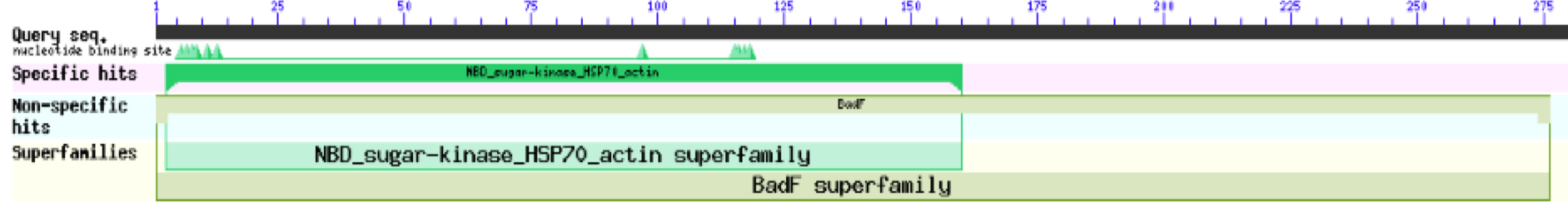
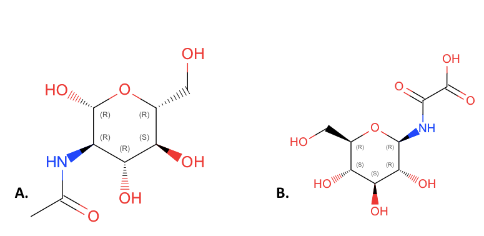
ZBS is a novel protein whose structure is solved but the function is unknown. This research was designed to attempt to uncover the function. Computational research indicated that N-acetylglucosamine (NAG) may be the potential substrate, and that the protein may phosphorylate NAG. This was determined using multiple computational tools, such as BLAST-P, DALI, SPRITE, InterPro. Molecular docking using NAG as a substrate was done with PyMol and Vina docking. After the computational research was completed, the protein was over-expressed and purified. The protein was used to test for activity with the substrate NAG. The kinase assay concluded that NAG is most likely not the substrate for 1ZBS due to a lack of specific activity.
Introduction
| |||||||||||
References
Proteopedia Page Contributors and Editors (what is this?)
Danielle Selover, Carmen Almendarez Rodriguez, Bonnie Hall, Jaime Prilusky