1ZBS is a protein with unknown function discovered through a genomics project of predictive folding. The goal of this project was to determine what the function of protein 1ZBS was using a combination of computational analysis and physical lab work.
For this project, we started by doing our computational work, which includes BLAST, DALI, SPRITE, Net-GO, and the Docking analysis, while we started transforming our bacteria, and over-expressing and purifying our protein for the kinase assays. From the computational analysis, it was believed that 1ZBS could be a NAGK protein. Further analysis into the kinase activity did not fully support this conclusion, as the specific activity was too low.
Sequence Alignments
BLAST-P
The first type of analysis that was done was using Protein Blast from NCBI. The analysis was first done as it only gave very broad information about the potential function of the protein, by matching domains across other proteins with known functions.
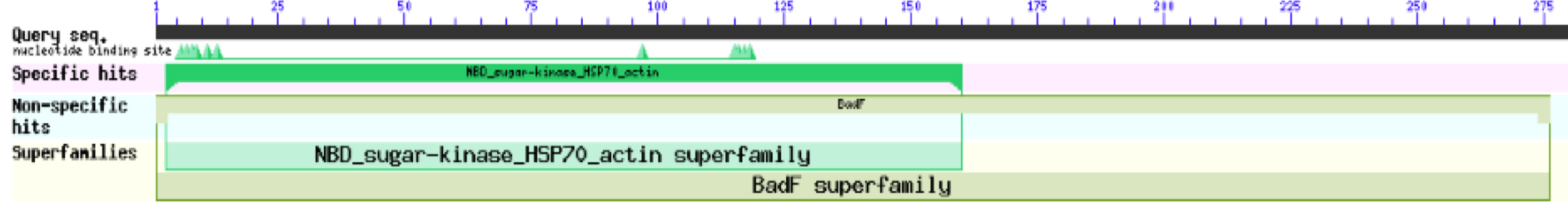 These results for 1ZBS showed that it was a match to the superfamily BADF. BADF is the superfamily that NAGK proteins also belong in so this was one of the reasons that we believed that 1ZBS may potentially be a NAGK protein.
These results for 1ZBS showed that it was a match to the superfamily BADF. BADF is the superfamily that NAGK proteins also belong in so this was one of the reasons that we believed that 1ZBS may potentially be a NAGK protein.
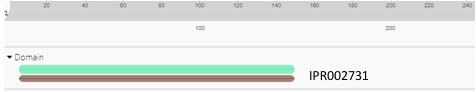
InterPro
InterPro is another sequence alignment software that we used to analyze the protein 1ZBS. This software gave similar results to. the Blast-P analysis, yet again giving matches to the BadF superfamily. The results showed a match to the general superfamily (green), as well as a match to the ATPase domain of the BadF family. These results further supported the idea that our protein should belong to this superfamily. On it's own, this is helpful for a starting place to determine the function but given the rest of the data from the analysis, it helps solidify the potential function of the 1ZBS protein.
Structural Alignments
DALI
DALI is a global alignment software that is used to match proteins up to possible structural matches within the Protein Data Bank (PDB).

This is the DALI alignment of 1ZBS and 2CH5, where 1ZBS is colored in green and is colored in yellow. The best results from the DALI alignment were mainly proteins with unknown functions, which makes it difficult to deduce a possible function off of. Because of these results, 2CH5, while only matching parts of the sequence was the best result for this analysis as it's function is known. The function of 2CH5 is a NAG kinase, which would further support the thought that 1ZBS could also be a NAG kinase.
Additional Tools =
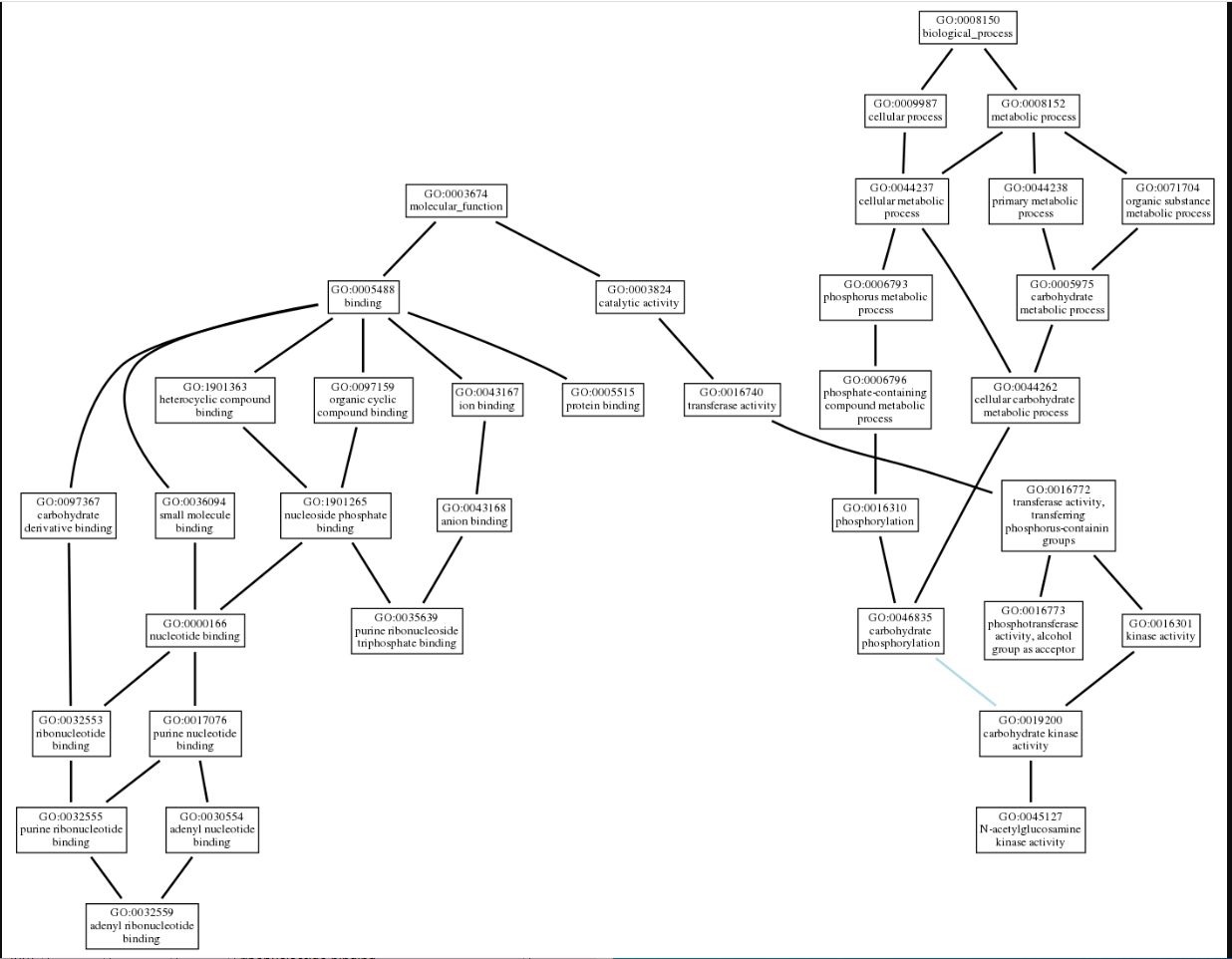
NetGO
We used NetGo 2.0 to help visualize the potential function of 1ZBS. This software uses predictive modeling to determine potential functions of the protein in query, and produces a set of gene ontology(GO) terms that best aligns with the protein's possible function. It starts with the most general information and builds to become more specific. These results came back in a list form. We then used AmiGo to visualize the connection between the terms. The final results landed us with a protein that has a purine ribonucleotide triphosphate binding domain, and could have some NAG kinase activity.
Docking Analysis
The docking analysis was started by using the software POCASA to help visualize the potential binding pockets in 1ZBS, as there was no literature available. These results, as shown below, helped us to better determine where we would limit the binding area to in PyRx.
From there we used PyRx to do molecular docking into the 1ZBS protein. This was completed using various different substrates in combination with ATP, which is needed for the kinase to be able to function. The control used was Imidazole which had a binding affinity of -2.9 kcal/mol. By completing this first, we had a better idea of what our binding affinity should be. Then we used to better determine if it was our substrate, as well as other similar structures found using the PDB. NAG had a binding affinity of -5.54kcal/mol. This was a better result than our control, and this helped further the belief that 1ZBS could be a NAG kinase.
The docking results did also give us a substrate with much more desirable binding affinity, which was N-(carboxycarbonyl)-glucosylamine, otherwise referred to as . This substrate had a binding affinity of -6.08 kcal/mol.
Substrate Possibilities
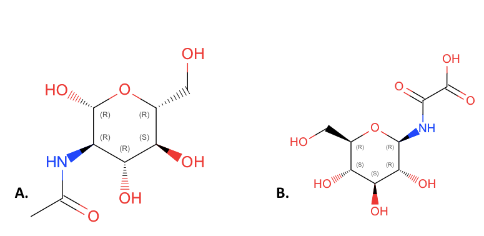
This figure compares the structures of NAG(A) and 4GP(B). All of the components of NAG are present in 4GP but they have been rearranged and an additional carbonyl group has been added to 4GP. We choose NAG as our substrate over 4GP because it is an inhibitor. Due to this, it is expected that 4GP will have better binding as it will bind to the transition state of the enzyme, however it will stop the function of the enzyme. This fact made it more ideal to test NAG over 4GP because NAG will give insight into the actual function of the substrate.
Kinase Assays
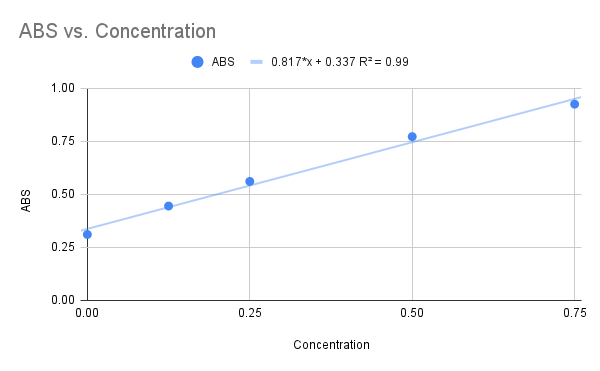
We used NAG as our final substrate for the 1ZBS kinase assay, but before the kinase assay could be run, a Bradford Assay was needed to determine the concentration of the 1ZBS protein that was over-expressed and purified in the lab. The Bradford Assay gave us the graph below with an R squared value of 0.99, and the equation listed on the graph.
Conclusion
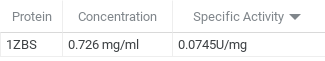
The goal of this experiment was to determine the function of 1zbs. We did this by creating a standard curve of BSA to compare our curve to our possible substrate NAG. This was done by a series of dilutions and Bradford assays to obtain a dilution factor by comparing it the absorbance levels of BSA. We concluded from our kinase assay results that 1ZBS is not a kinase because the specific activity was 0.074 U/mg.








