BASIL2023GVP76586
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Molecular Docking == | == Molecular Docking == | ||
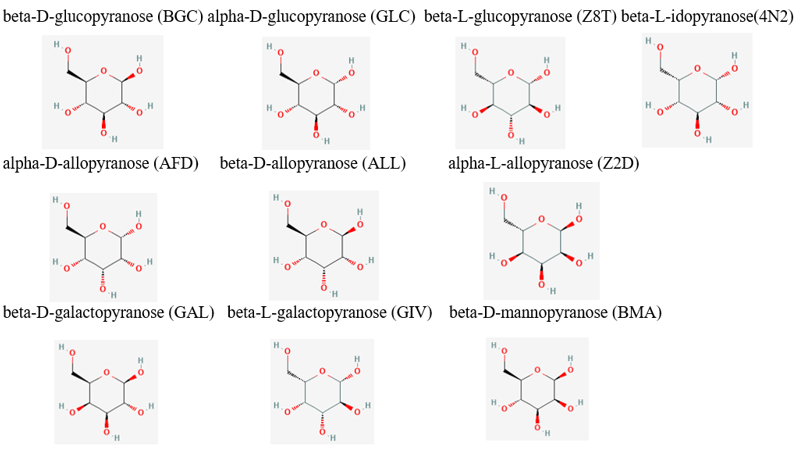
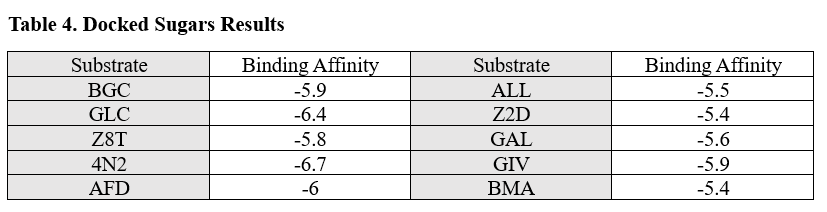
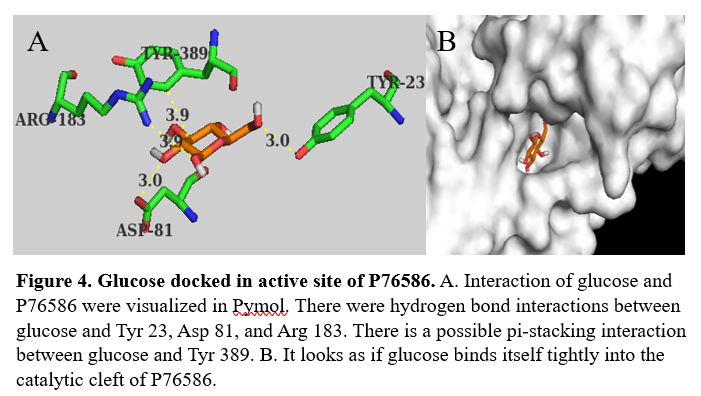
From the initial search throughout all of the computational tools, we decided that our putative kinase was potentially a glucokinase. We docked other sugars along with glucose in figure 1. | From the initial search throughout all of the computational tools, we decided that our putative kinase was potentially a glucokinase. We docked other sugars along with glucose in figure 1. | ||
| + | |||
[[Image:Sugar_substrates.png]] | [[Image:Sugar_substrates.png]] | ||
| Line 30: | Line 31: | ||
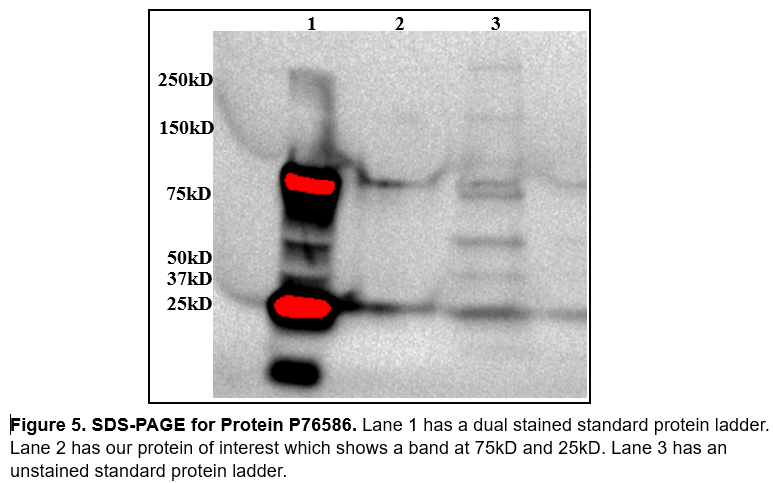
Our protein of interest has a weight of ≈44.53kD. When analyzing SDS PAGE (figure 5) we were slightly concerned we weren't working with our protein of interest. We didn't get a great image out of SDS, if there were more time we would run again with more protein in the well so that we could see it better. We also made the mistake of not including our induction samples and our fractions from protein purification using nickle affinity chromatography. | Our protein of interest has a weight of ≈44.53kD. When analyzing SDS PAGE (figure 5) we were slightly concerned we weren't working with our protein of interest. We didn't get a great image out of SDS, if there were more time we would run again with more protein in the well so that we could see it better. We also made the mistake of not including our induction samples and our fractions from protein purification using nickle affinity chromatography. | ||
[[Image:SDS.png]] | [[Image:SDS.png]] | ||
| + | |||
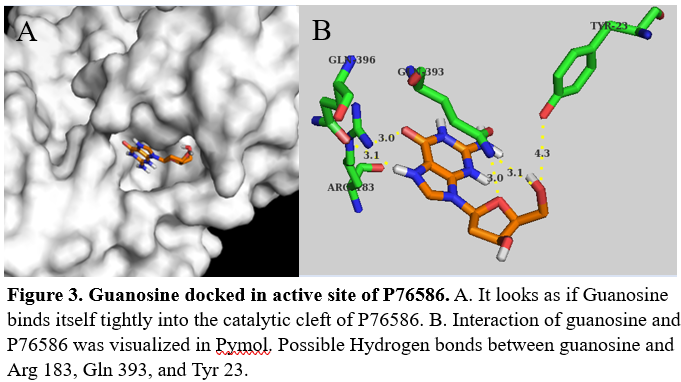
We ran two coupled kinase assays, one with glucose and one with guanosine. After calculating the specific activity of our protein with glucose and guanosine we determined there was not enough activity for glucose or guanosine to be the substrate. | We ran two coupled kinase assays, one with glucose and one with guanosine. After calculating the specific activity of our protein with glucose and guanosine we determined there was not enough activity for glucose or guanosine to be the substrate. | ||
Revision as of 18:25, 24 April 2023
Investigating the Function of Protein P76586
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644