This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Anum-II
From Proteopedia
| Line 8: | Line 8: | ||
| - | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (His48,Tyr52, Tyr73 and Asp99) except for Asp49 which is replaced by Lys 49. | + | Anum-II has a length of 134 amino-acids. The phospholipase is formed by a short N-terminalαhelix (between residues 2-12), a 2nd αhelix (residues 40-55), two-stranded antiparallel <scene name='Sandbox136/Feuillets/1'>sheet</scene> linked thanks to a βwing (74-85) and a 3rd αhelix (residues 90-107). The 3rd αhelix is bound to the 2nd αhelix (in an antiparallele way) thanks to <scene name='Sandbox136/Disulfide/1'>disulfide bonds </scene> ([Cys 44-Cys 105] and [Cys 51-Cys 98]) and thus form a <scene name='Sandbox136/Stabilisation/1'>rigid platform</scene>. The protein is stabilized by 5 other disulfides bonds [Cys 27-Cys 125], [Cys 29-Cys 45] [Cys 50 Cys 134] [Cys 61-Cys 91] [Cys84-Cys96]. Alignment of Anum-II with other PLA2 have revealed that the positions of amino-acid residues which form the <scene name='Sandbox136/Site_catalytique/2'>catalityc apparatus</scene> are conserved (<scene name='49/497100/Active_site_residues_with_fas/2'>His48,Tyr52, Tyr73 and Asp99</scene>) except for Asp49 which is replaced by Lys 49. |
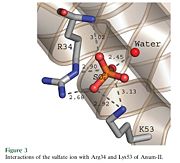
The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | The structure of the protein has revealed the presence of an anion-binding site (Murakami ''et al.'', 2006) between <scene name='Sandbox136/Anion/2'>R34</scene>, <scene name='Sandbox136/Anion/2'>K53</scene> and a water molecule (see Figure 3). Sulfate ion is anchored thanks to hydrogen bonds between: | ||
Current revision
| |||||||||||
Bibliography
Holland, D.R., Clancy, L.L., Muchmore, S.W., Ryde, T.J., Einspahr, H.M., Finzel, B.C., Heinrikson, R.L., Watenpaugh, K.D. (1990).Biochem-J .265,17649-56
Chioato, L., De Oliveira, A. H., Ruller, R., Sa., J. M., Ward, R. J. (2002).Biochem-J .366,971-6
Murakami, M. T. , Melo,C. C., Angulo,Y. , Lomonte,B. Arni, R. K. (2006).Acta Cryst.F62, 423-426
Yamazaki, Y., Matsunaga, Y., Nakano, Y. & Morita, T. (2005). J. Biol. Chem. 280, 29989–29992.
Created with the participation of Elie Peillon