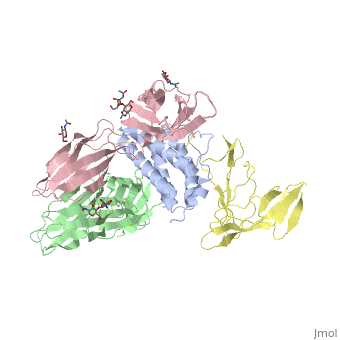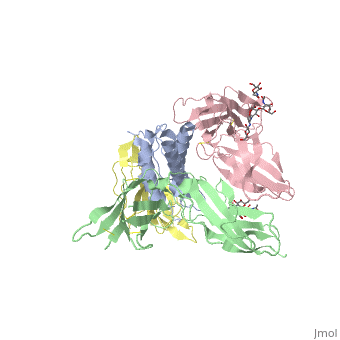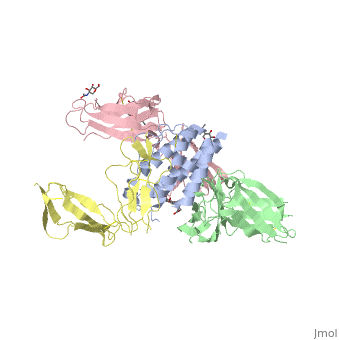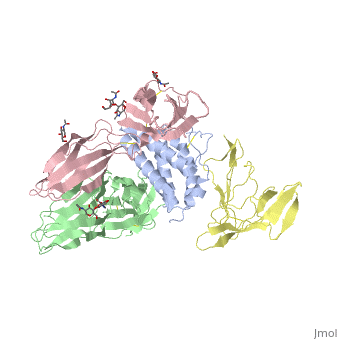We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Interleukin receptor
From Proteopedia
(Difference between revisions)
| Line 6: | Line 6: | ||
See also: [[Cytokine receptors]]. | See also: [[Cytokine receptors]]. | ||
| - | '''TYPE-1 INTERLEUKIN-1 RECEPTOR COMPLEXED WITH INTERLEUKIN-1 BETA''' | + | =='''TYPE-1 INTERLEUKIN-1 RECEPTOR COMPLEXED WITH INTERLEUKIN-1 BETA''' == |
[http://en.wikipedia.org/wiki/Interleukin-1_receptor Interleukin-1 receptor] complex with ligand and go through the plasma membrane. <scene name='57/571319/Scene_2/2'> Type 1 Interleukin-1 receptor complex with Interleukin-1 beta</scene> 3D structure is showing here. Ribbon diagram of s-IL 1R complex to IL-1β. The complex has approximate dimensions of 97Å × 52Å ×35Å with one s-IL1R molecule wrapping around the IL-1β molecule with 1:1 ratio. In the <scene name='57/571319/Scene_2/3'>complex</scene>, domain 3 provides a 'lid' which covers most of the top of the IL-1β β-barrel, whereas domains 1 and 2 from a groove which binds to the lower rim of the barrel. Here, Domains 1,2 and 3 of s-IL 1R are colored light, medium and dark blue, respectively. IL-1β is yellow, with site A residues in green and site B residues in red. The structure is oriented so that the carboxy terminus of s-IL 1R and the cell membrane are at the bottom of the picture. | [http://en.wikipedia.org/wiki/Interleukin-1_receptor Interleukin-1 receptor] complex with ligand and go through the plasma membrane. <scene name='57/571319/Scene_2/2'> Type 1 Interleukin-1 receptor complex with Interleukin-1 beta</scene> 3D structure is showing here. Ribbon diagram of s-IL 1R complex to IL-1β. The complex has approximate dimensions of 97Å × 52Å ×35Å with one s-IL1R molecule wrapping around the IL-1β molecule with 1:1 ratio. In the <scene name='57/571319/Scene_2/3'>complex</scene>, domain 3 provides a 'lid' which covers most of the top of the IL-1β β-barrel, whereas domains 1 and 2 from a groove which binds to the lower rim of the barrel. Here, Domains 1,2 and 3 of s-IL 1R are colored light, medium and dark blue, respectively. IL-1β is yellow, with site A residues in green and site B residues in red. The structure is oriented so that the carboxy terminus of s-IL 1R and the cell membrane are at the bottom of the picture. | ||
| - | '''STRUCTURE OF THE INTERLEUKIN-1BETA SIGNALING COMPLEX''' | + | =='''STRUCTURE OF THE INTERLEUKIN-1BETA SIGNALING COMPLEX'''== |
[[Image:Ternary complex paradigm.png||250px|right|]] | [[Image:Ternary complex paradigm.png||250px|right|]] | ||
Revision as of 10:08, 4 June 2023
| |||||||||||