We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Valentina Dutton/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 7: | Line 7: | ||
The vast majority of mitochondrial proteins have an N-terminal sequence that indicates that they must be exported to the mitochondria. This sequence is called the mitochondrial signal peptide. When the protein enters the mitochondria, this sequence is usually cleaved, providing greater stability. It is still unknown what this sequence would be in the case of PGAM5 since the protein signal peptide is not cleaved<ref name="sie">PMID:35921890</ref> and PGAM5 is anchored in its entirety to the inner membrane through its transmembrane domain, defined by amino acids 9-29<ref name="cha">PMID:28648608</ref>. | The vast majority of mitochondrial proteins have an N-terminal sequence that indicates that they must be exported to the mitochondria. This sequence is called the mitochondrial signal peptide. When the protein enters the mitochondria, this sequence is usually cleaved, providing greater stability. It is still unknown what this sequence would be in the case of PGAM5 since the protein signal peptide is not cleaved<ref name="sie">PMID:35921890</ref> and PGAM5 is anchored in its entirety to the inner membrane through its transmembrane domain, defined by amino acids 9-29<ref name="cha">PMID:28648608</ref>. | ||
| - | Regarding the catalytic activity, histidine 105 is responsible for the nucleophilic attack of the phosphate of the target protein, performing the intermediate link between the protein and the phosphate. However, histidine 105 is part of the canonical RHGE motif, present in all proteins of the PGAM family, forming part of the PGAM domain (98-289)<ref name="cha" />. The WDXNWD motif at amino acids 58-63 has been shown to function as an allosteric regulator of the specific phosphatase activity of PGAM5 by inducing protein oligomerization<ref name="wil" />. Mutations in this motif prevent oligomerization of the enzyme but still present as dimers since the C-terminal tail (270-289) in PGAM5 is responsible for the dimerization of the protein<ref name="cha" />. These dimers, however, do not show phosphatase activity<ref name="wil" />. | + | Regarding the catalytic activity, <scene name='96/969636/His105/1'>histidine 105</scene> is responsible for the nucleophilic attack of the phosphate of the target protein, performing the intermediate link between the protein and the phosphate. However, histidine 105 is part of the canonical RHGE motif, present in all proteins of the PGAM family, forming part of the PGAM domain (98-289)<ref name="cha" />. The WDXNWD motif at amino acids 58-63 has been shown to function as an allosteric regulator of the specific phosphatase activity of PGAM5 by inducing protein oligomerization<ref name="wil" />. Mutations in this motif prevent oligomerization of the enzyme but still present as dimers since the C-terminal tail (270-289) in PGAM5 is responsible for the dimerization of the protein<ref name="cha" />. These dimers, however, do not show phosphatase activity<ref name="wil" />. |
== Function == | == Function == | ||
Even though PGAM5 is a member of the PGAM protein family, it appears to lack phosphoglycerate mutase typical phosphotransferase and/or phosphohydrolase activities.<ref name="cha" />(UniProt). Instead, this protein is a serine/threonine (Ser/Thr) phosphatase, that is, it’s responsible for protein-protein interactions through dephosphorylation of serine/threonine and, occasionally, histidine residues<ref name="che">PMID:33370650</ref><ref name="shi">PMID:19879837</ref>. Its active site is composed of a histidine residue (His-105) responsible for the nucleophilic attack of the phosphorus atom acting as a phospho-acceptor<ref name="cha" /><ref name="shi" />. | Even though PGAM5 is a member of the PGAM protein family, it appears to lack phosphoglycerate mutase typical phosphotransferase and/or phosphohydrolase activities.<ref name="cha" />(UniProt). Instead, this protein is a serine/threonine (Ser/Thr) phosphatase, that is, it’s responsible for protein-protein interactions through dephosphorylation of serine/threonine and, occasionally, histidine residues<ref name="che">PMID:33370650</ref><ref name="shi">PMID:19879837</ref>. Its active site is composed of a histidine residue (His-105) responsible for the nucleophilic attack of the phosphorus atom acting as a phospho-acceptor<ref name="cha" /><ref name="shi" />. | ||
| Line 16: | Line 16: | ||
Additionally, it’s been suggested that PGAM5 can also be involved in cellular senescence, necroptosis, responses to stress, parasite replication, lipid metabolism, inflammatory and immune responses<ref name="bab" />. | Additionally, it’s been suggested that PGAM5 can also be involved in cellular senescence, necroptosis, responses to stress, parasite replication, lipid metabolism, inflammatory and immune responses<ref name="bab" />. | ||
| - | == Subcelluar | + | == Subcelluar Localization == |
PGAM5 is a mitochondrial enzyme. However, the mitochondria is a complex organelle, composed of really distinct regions. It has 2 membranes, an outer membrane (OMM), in direct contact with the cytoplasm, and an inner membrane (IMM), in contact with the mitochondrial matrix. Between them there is the intermembrane space, where the protons from the matrix are directed Therefore, the mitochondrial matrix exhibits a higher pH than the intermembrane space. | PGAM5 is a mitochondrial enzyme. However, the mitochondria is a complex organelle, composed of really distinct regions. It has 2 membranes, an outer membrane (OMM), in direct contact with the cytoplasm, and an inner membrane (IMM), in contact with the mitochondrial matrix. Between them there is the intermembrane space, where the protons from the matrix are directed Therefore, the mitochondrial matrix exhibits a higher pH than the intermembrane space. | ||
| Line 48: | Line 48: | ||
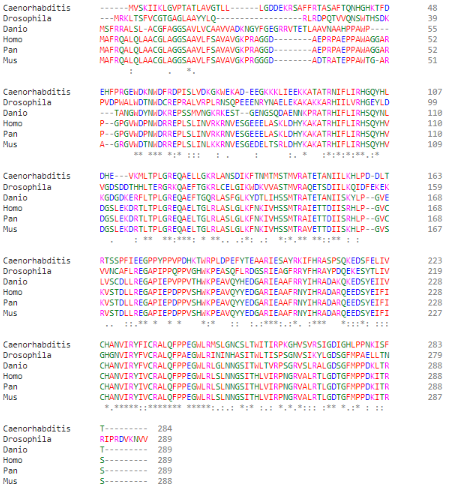
(Highlight of the His-105 of PGAM5 in different species, them being: (top to bottom) ''C. elegans; D. melanogaster; D. rerio; H. sapiens; P. paniscus; M. musculus'') | (Highlight of the His-105 of PGAM5 in different species, them being: (top to bottom) ''C. elegans; D. melanogaster; D. rerio; H. sapiens; P. paniscus; M. musculus'') | ||
| - | == Related | + | == Related Diseases == |
It’s suggested that PGAM5 might possibly be associated with diseases caused by mitochondrial dysfunction, especially if it occurs in response to defective mitophagy process, including neurodegenerative diseases<ref name="lia">PMID:34630036</ref><ref name="lue">PMID:25222142</ref>. | It’s suggested that PGAM5 might possibly be associated with diseases caused by mitochondrial dysfunction, especially if it occurs in response to defective mitophagy process, including neurodegenerative diseases<ref name="lia">PMID:34630036</ref><ref name="lue">PMID:25222142</ref>. | ||
Revision as of 00:55, 22 June 2023
Human Phosphoglycerate Mutase Family Member 5 (PGAM5)
| |||||||||||
References
- ↑ 1.0 1.1 1.2 1.3 Takeda K, Komuro Y, Hayakawa T, Oguchi H, Ishida Y, Murakami S, Noguchi T, Kinoshita H, Sekine Y, Iemura S, Natsume T, Ichijo H. Mitochondrial phosphoglycerate mutase 5 uses alternate catalytic activity as a protein serine/threonine phosphatase to activate ASK1. Proc Natl Acad Sci U S A. 2009 Jul 28;106(30):12301-5. Epub 2009 Jul 9. PMID:19590015 doi:http://dx.doi.org/0901823106
- ↑ 2.0 2.1 2.2 Wilkins JM, McConnell C, Tipton PA, Hannink M. A conserved motif mediates both multimer formation and allosteric activation of phosphoglycerate mutase 5. J Biol Chem. 2014 Sep 5;289(36):25137-48. PMID:25012655 doi:10.1074/jbc.M114.565549
- ↑ 3.0 3.1 Siebert V, Silber M, Heuten E, Muhle-Goll C, Lemberg MK. Cleavage of mitochondrial homeostasis regulator PGAM5 by the intramembrane protease PARL is governed by transmembrane helix dynamics and oligomeric state. J Biol Chem. 2022 Jul 31:102321. doi: 10.1016/j.jbc.2022.102321. PMID:35921890 doi:http://dx.doi.org/10.1016/j.jbc.2022.102321
- ↑ 4.0 4.1 4.2 4.3 4.4 Chaikuad A, Filippakopoulos P, Marcsisin SR, Picaud S, Schroder M, Sekine S, Ichijo H, Engen JR, Takeda K, Knapp S. Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly. Structure. 2017 Jul 5;25(7):1089-1099.e3. doi: 10.1016/j.str.2017.05.020. Epub, 2017 Jun 22. PMID:28648608 doi:http://dx.doi.org/10.1016/j.str.2017.05.020
- ↑ Cheng M, Lin N, Dong D, Ma J, Su J, Sun L. PGAM5: A crucial role in mitochondrial dynamics and programmed cell death. Eur J Cell Biol. 2021 Jan;100(1):151144. PMID:33370650 doi:10.1016/j.ejcb.2020.151144
- ↑ 6.0 6.1 Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009 Oct 30;139(3):468-84. PMID:19879837 doi:10.1016/j.cell.2009.10.006
- ↑ 7.0 7.1 Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem. 2006 Dec 8;281(49):37893-903. PMID:17046835 doi:10.1074/jbc.M606539200
- ↑ Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008 May 1;314(8):1789-803. doi: 10.1016/j.yexcr.2008.02.014. Epub , 2008 Mar 5. PMID:18387606 doi:http://dx.doi.org/10.1016/j.yexcr.2008.02.014
- ↑ 9.0 9.1 9.2 Baba T, Tanimura S, Yamaguchi A, Horikawa K, Yokozeki M, Hachiya S, Iemura SI, Natsume T, Matsuda N, Takeda K. Cleaved PGAM5 dephosphorylates nuclear serine/arginine-rich proteins during mitophagy. Biochim Biophys Acta Mol Cell Res. 2021 Jun;1868(7):119045. PMID:33872670 doi:10.1016/j.bbamcr.2021.119045
- ↑ 10.0 10.1 Liang MZ, Ke TL, Chen L. Mitochondrial Protein PGAM5 Emerges as a New Regulator in Neurological Diseases. Front Mol Neurosci. 2021 Sep 23;14:730604. PMID:34630036 doi:10.3389/fnmol.2021.730604
- ↑ 11.0 11.1 11.2 Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, Zhang Y, Dawson VL, Dawson TM, Lenardo M. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson's-like movement disorder. Nat Commun. 2014 Sep 15;5:4930. PMID:25222142 doi:10.1038/ncomms5930
- ↑ Ng Kee Kwong F, Nicholson AG, Pavlidis S, Adcock IM, Chung KF. PGAM5 expression and macrophage signatures in non-small cell lung cancer associated with chronic obstructive pulmonary disease (COPD). BMC Cancer. 2018 Dec 10;18(1):1238. PMID:30526542 doi:10.1186/s12885-018-5140-9
- ↑ He GW, Günther C, Kremer AE, Thonn V, Amann K, Poremba C, Neurath MF, Wirtz S, Becker C. PGAM5-mediated programmed necrosis of hepatocytes drives acute liver injury. Gut. 2017 Apr;66(4):716-723. PMID:27566130 doi:10.1136/gutjnl-2015-311247
- ↑ Li CJ, Lin LT, Tsai HW, Wen ZH, Tsui KH. Phosphoglycerate mutase family member 5 maintains oocyte quality via mitochondrial dynamic rearrangement during aging. Aging Cell. 2022 Feb;21(2):e13546. PMID:34995407 doi:10.1111/acel.13546