User:Nane Milene Sposito Almeida Pereira/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 10: | Line 10: | ||
'''General Aspects''' | '''General Aspects''' | ||
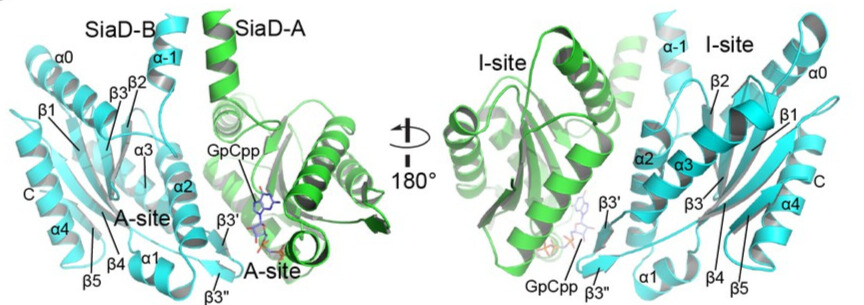
| - | The SiaC-SiaD complex is formed by <scene name='97/973098/Siad/1'>two SiaD molecules</scene> and the binding of <scene name='97/973098/Siac/1'>four SiaC</scene>. The conformation of the <scene name='97/973098/Complex/1'>complex</scene> is composed of two SiaD molecules (<font color="MediumSeaGreen">SiaD-A</font> and <font color="RoyalBlue">SiaD-B</font>) that form a parallel spiral through their helix rods, and their dimeric rods are stabilized by the binding of two pairs of SiaC molecules (<font color="Violet">SiaC-C</font>/<font color="yellow">SiaC-D</font> and <font color="MediumOrchid">SiaC-E</font>/<font color="SkyBlue">SiaC-F</font>), each in a different location along the dimeric stem, which may be proximal or distal to the DGC domain of SiaD. Furthermore, a non-hydrolyzable GTP analog molecule, <scene name='97/973098/Gpcpp/1'>GpCpp</scene>, was observed to bind to the active site of SiaD-A. | + | The SiaC-SiaD complex is formed by <scene name='97/973098/Siad/1'>two SiaD molecules</scene> and the binding of <scene name='97/973098/Siac/1'>four SiaC</scene>. The conformation of the <scene name='97/973098/Complex/1'>complex</scene> is composed of two SiaD molecules (<font color="MediumSeaGreen">SiaD-A</font> and <font color="RoyalBlue">SiaD-B</font>) that form a parallel spiral through their helix rods, and their dimeric rods are stabilized by the binding of two pairs of SiaC molecules (<font color="Violet">SiaC-C</font>/<font color="yellow">SiaC-D</font> and <font color="MediumOrchid">SiaC-E</font>/<font color="SkyBlue">SiaC-F</font>), each in a different location along the dimeric stem, which may be proximal or distal to the DGC domain of SiaD. Furthermore, a non-hydrolyzable GTP analog molecule, <scene name='97/973098/Gpcpp/1'>GpCpp</scene>, was observed to bind to the active site of SiaD-A. |
| + | |||
| + | The crystal structure of full-lenght SiaD in complex with SiaC was determined by integrating the diffraction data and scaling them using the program HKL3000 to space group C2221 at 2.65 Å resolution, and the structure was subsequently determined through molecular replacement using the published conserved DGC domain of the [[WspR]] structure (PDB code: 3BRE) and the SiaC structure (PDB code: 6KKP). | ||
'''The DGC domain of SiaD''' | '''The DGC domain of SiaD''' | ||
Revision as of 02:08, 22 June 2023
SiaC-SiaD Complex
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Chen G, Zhou J, Zuo Y, Huo W, Peng J, Li M, Zhang Y, Wang T, Zhang L, Zhang L, Liang H. Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa. Elife. 2021 Sep 9;10:e67289. PMID:34498587 doi:10.7554/eLife.67289