User:Nane Milene Sposito Almeida Pereira/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 16: | Line 16: | ||
'''The DGC domain of SiaD''' | '''The DGC domain of SiaD''' | ||
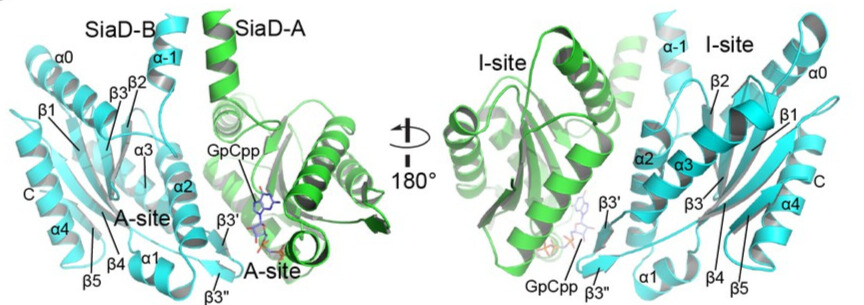
| - | The C-terminal GGDEF domain of SiaD is highly conserved. Two DGC domains in the complex are oriented anti-parallel, with their active sites facing each other. Each DGC domain consists of a core canonical fold of five antiparallel β-strands (β1–β5) around which five α-helices (α0–α4) and two short antiparallel β-strands are wrapped (β3ʹ and β3"), forming a highly conserved GTP substrate-binding site (GGDEF domain, A-site) and a c-di-GMP product binding/inhibitory site (I-site). | + | The C-terminal GGDEF domain of SiaD is highly conserved. Two DGC domains in the complex are oriented anti-parallel, with their active sites facing each other. Each DGC domain consists of a core canonical fold of five antiparallel β-strands (β1–β5) around which five α-helices (α0–α4) and two short antiparallel β-strands are wrapped (β3ʹ and β3"), forming a highly conserved GTP substrate-binding site (GGDEF domain, A-site) and a c-di-GMP product binding/inhibitory site (I-site). Furthermore, residues involved in GTP and Mg 2+ binding have been shown to be essential for the DGC activity of SiaD. |
[[Image:Overview_of_the_DGC_domain_dimer.jpg]] | [[Image:Overview_of_the_DGC_domain_dimer.jpg]] | ||
Revision as of 02:32, 22 June 2023
SiaC-SiaD Complex
| |||||||||||
References
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644
- ↑ Chen G, Zhou J, Zuo Y, Huo W, Peng J, Li M, Zhang Y, Wang T, Zhang L, Zhang L, Liang H. Structural basis for diguanylate cyclase activation by its binding partner in Pseudomonas aeruginosa. Elife. 2021 Sep 9;10:e67289. PMID:34498587 doi:10.7554/eLife.67289