Function and Structural highlights
The structure of the NF-Y protein described here was generated by a crystallography using synchrotron radiation at 3.1Å resolution. The molecule consists of a heterotrimer complexed with a 25bp DNA fragment containing a CCAAT sequence.
NF-Y is a highly conserved (strongly conserved in all eukaryotes) and ubiquitously expressed protein that acts as a transcription factor, binding directly to a specific site on the DNA, the CCAAT box, a sequence that can be found in the promoter of many protein coding genes. Studies show that the CCAAT sites in the genome are located upstream of the TSS site in the promoter of those genes. This protein is composed of 3 subunits, NF-YA (subunit Alpha), NF-YB (subunit Beta) and NF-YC (subunit Gamma), which are all necessary for DNA binding. The NF-YB and NF-YC subunits host histone fold-domains (HFDs), which are present in the core of histones and allow binding to the DNA. NF-YB and NF-YC show homology in sequence with histones H2A and H2B, respectively, hence, the Beta and Gamma subunits can bind directly to the DNA, but require NF-YA association to stabilize the binding of the complex to the DNA.
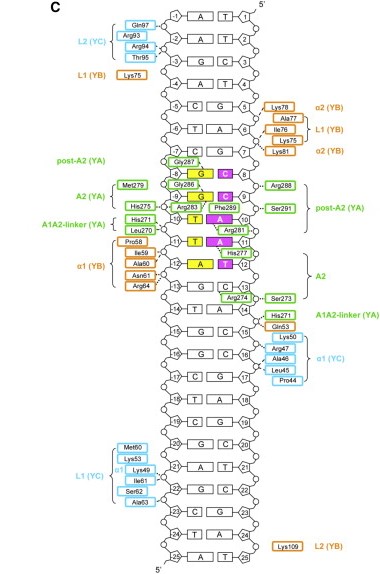
The heterodimerization of NF-YB and NF-YC allow NF-YA to associate with the complex, through binding of its A1 helix to NF-YB and NF-YC HFD domains, then allowing binding to CCAAT. NF-YA has a specific C-terminal domain responsible for CCAAT recognition, its A2 helix, which searches for the DNA motif for binding, but all of the 3 subunits can contact DNA after CCAAT binding. The contact regions for NF-Y subunits are shown in the image below.
After finding the motif, NF-YA inserts its A2 helix into the DNA minor groove, which causes a bend in the DNA, allowing, then, other transcriptional factors to bind in adjacent DNA grooves. The binding of NF-YA is shown in the image below.
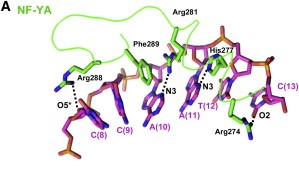
It was hypothesized that NF-YA is recruited by the HFD domain because it would function as a histone tail to allow and stabilize DNA binding with the NF-Y complex. Indeed, the core structures the histone dimer H2A/H2B overlay with NF-YB and NF-YC, while NF-YA overlay with the HB2 tail, indicating a similar function for those structures.
Therefore, the main function of NF-Y is related to its high specificity and affinity binding to DNA, which induces gene promoter activation with the help of other transcriptional factors, although its binding can also induce posttranslational alterations in histones, such as methylations and acetylations, being also capable of establishing acetylation marks directly on gene promoters through recruitment of enzymes responsible for inducing those alterations. This epigenetic marks can induce or repress gene transcription by modifying chromatin accessibility. Hence, NF-Y can primary bind into a promoter region where transcription preinitiation complexes can be established, avoiding nucleosome formation and giving space for other transcription initiation proteins (some of those can interact directly with NF-Y and hence be recruited by it) and TFs to bind to the promoter of a gene that will be transcripted. It is hypothesized that, since NF-Y have a similar structure to histones, its binding to the DNA results in incompatibility for other histones binding to the same region. This primary binding also can recruit histone modifying enzymes, which regulate chromatin accessibility. Another possible function for NF-Y is that it can ensure the usage of the correct TSS site for transcription. It was shown that NF-YA depletion in a cell can cause histone binding to the correct TSS, which result in the relocation of the preinitiation complex to another TSS that is upstream of the canonical one. This shift in the position of the TSS can result in a longer mRNA that could be originated from another ORF, resulting in abnormal mRNAs. This function of NF-Y could be explained by the fact that this TF predominantly binds to CG-rich promoters of essential genes (such as those related to DNA repair, cell cycle and transcription regulation), which need to be transcripted correctly.
Here we show the molecule structure while binding to the DNA, with each subunit represented in a different color: NF-YA in blue, NF-YB in green and NF-YC in pink, while the DNA is represented in grey. Here is its and its with alpha-helixes colored in pink, and beta-sheets colored in yellow. Using the representation we can see the secondary structures in each subunit.
NF-Y and diseases
It was shown that NF-Y binding sites are abundant in promoters of genes that induce cell growth or that are responsible for cell transformation. Therefore, NF-Y could induce transcription of oncogenes, leading to cell growth and transformation in normal cells, which can result in tumor-like cells. Overexpression of NF-Y subunits was already described in different types of cancers such as lung, breast, thyroid and prostate cancer.
NF-Y subunits like NF-YA and NF-YC can form aggregates with mutant huntingtin (HTT) protein, which is abundant in neurons of patients with Hungtinton’s disease. By sequestering NF-Y subunits, mutant HTT impairs NF-Y dependent transcriptional activation of genes, such as HSP70, that encodes for a chaperone protein responsible for degradation of mutant HTT aggregates. In conclusion, NF-Y is required for adequate transcription of chaperones, and its absence in the CCAAT sites can induce aggregates accumulation, which aggravates the disease.
This molecule can also be involved in diabetes, when its DNA binding in promoter of enzymes related to glucose metabolism is either reduced or augmented, causing non ideal transcription of the downstream genes, which impact in regulation of blood glucose levels.
Other circumstances where mutations of the NF-Y binding site occur can result in the block of NF-Y binding, altering the expression of genes regulated by NF-Y transcriptional regulation.


