User:Marcos Vinícius Caetano/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 8: | Line 8: | ||
[[Image:MyoVI.png|600px]] | [[Image:MyoVI.png|600px]] | ||
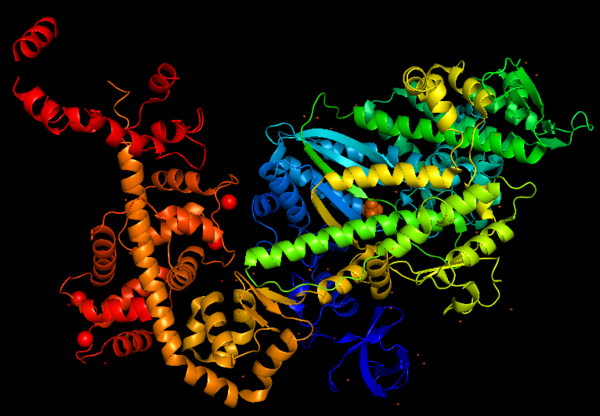
''Figure 1: Representation of Myosin VI structure in gradient rainbow representation, where <span style="color:blue">'''blue'''</span> is N-terminal (5’) and <span style="color:red">'''red'''</span> the C-terminal (3’).'' | ''Figure 1: Representation of Myosin VI structure in gradient rainbow representation, where <span style="color:blue">'''blue'''</span> is N-terminal (5’) and <span style="color:red">'''red'''</span> the C-terminal (3’).'' | ||
| + | |||
The '''basic structure''' of myosin VI, and most myosin molecule heavy chains, consists of '''three regions''': | The '''basic structure''' of myosin VI, and most myosin molecule heavy chains, consists of '''three regions''': | ||
| Line 75: | Line 76: | ||
*'''Function''': Redirectionare the lever arm and contains a new CaM-binding motif. | *'''Function''': Redirectionare the lever arm and contains a new CaM-binding motif. | ||
| - | *'''Mechanism''': The proximal part of insert 2 (<scene name='97/973101/774_812_distal_and_proximal/3'>Pro774-Trp787</scene> - in <span style="color:red">'''red'''</span>) wraps around the <scene name='97/973101/774_812_distal_and_proximal/4'>converter</scene> (in <span style="color:blue">'''blue'''</span>), while the distal part (<scene name='97/973101/774_812_distal_and_proximal/3'>Trp787-Tyr812</scene> - in <span style="color:green">'''green'''</span>) forms a CaM-binding motif. The insert 2 and its associated CaM molecule (with 4Ca<sup>2+</sup>), make specific interactions with the converter, many involving a variable loop (<scene name='97/973101/Insert_2_full/1'>Lys719-Pro731</scene>- in <span style="color:magenta">'''magenta'''</span>). In addiction, there is '''apolar interactions''' that stabilize the proximal part of insert 2 on the surface of the converter, where <scene name='97/973101/Insert_2_full/3'>hidrophofobic side chains</scene> from the amino acids <span style="color:gold">'''F763, F766, M770'''</span> stabilize the orientation of the last helix <scene name='97/973101/Insert_2_full/4'>(representation with cartoon and stick)</scene>. The result of <scene name='97/973101/Insert_2_full/2'>interactions</scene> is that <scene name='97/973101/Iq_helix_emerge/1'>IQ helix</scene> - in <span style="color:green">'''green'''</span>) '''emerges ~120°''' from the position that it emerges in all other myosins, '''redirecting the IQ helix''' and the CaM towards the '''minus-end''' of the actin filament. '''This is what makes myosin VI unique'''. | + | *'''Mechanism''': The proximal part of insert 2 (<scene name='97/973101/774_812_distal_and_proximal/3'>Pro774-Trp787</scene> - in <span style="color:red">'''red'''</span>) wraps around the <scene name='97/973101/774_812_distal_and_proximal/4'>converter</scene> (in <span style="color:blue">'''blue'''</span>), while the distal part (<scene name='97/973101/774_812_distal_and_proximal/3'>Trp787-Tyr812</scene> - in <span style="color:green">'''green'''</span>) forms a CaM-binding motif. The insert 2 and its associated CaM molecule (with 4Ca<sup>2+</sup>), make specific interactions with the converter, many involving a variable loop (<scene name='97/973101/Insert_2_full/1'>Lys719-Pro731</scene>- in <span style="color:magenta">'''magenta'''</span>). In addiction, there is '''apolar interactions''' that stabilize the proximal part of insert 2 on the surface of the converter, where <scene name='97/973101/Insert_2_full/3'>hidrophofobic side chains</scene> from the amino acids <span style="color:gold">'''F763, F766, M770'''</span> stabilize the orientation of the last helix <scene name='97/973101/Insert_2_full/4'>(representation with cartoon and stick)</scene>. Also, there is <scene name='97/973101/Insert_2_full_salt_bridges/2'>salt-bridge interactions</scene> between the converter and both lobes of CaM, throghout the amino acids: <span style="color:orange">''' D730, E114, K795, R792'''</span>; <span style="color:blue">'''K736, E14, R732'''</span>; and <span style="color:gold">'''R728, E120'''</span>. The result of <scene name='97/973101/Insert_2_full/2'>interactions</scene> is that <scene name='97/973101/Iq_helix_emerge/1'>IQ helix</scene> - in <span style="color:green">'''green'''</span>) '''emerges ~120°''' from the position that it emerges in all other myosins, '''redirecting the IQ helix''' and the CaM towards the '''minus-end''' of the actin filament. '''This is what makes myosin VI unique'''. |
The figure below compare the orientation of the last helix of the converter in myosin VI (<span style="color:green">'''green'''</span>) with myosin V (<span style="color:blue">'''blue'''</span>), and it shows a '''difference of 19°''', that combined with the apolar and salt-bridges interactions, makes the emerge of the IQ helix 120° from the position that it emerges in all other myosins. | The figure below compare the orientation of the last helix of the converter in myosin VI (<span style="color:green">'''green'''</span>) with myosin V (<span style="color:blue">'''blue'''</span>), and it shows a '''difference of 19°''', that combined with the apolar and salt-bridges interactions, makes the emerge of the IQ helix 120° from the position that it emerges in all other myosins. | ||
| Line 81: | Line 82: | ||
''Figure 4: Alignement of myosin VI structure (2BKI) (<span style="color:green">'''in green'''</span>) with myosin V (1OE9) (<span style="color:blue">'''blue'''</span>), focusing on the last helix to show the angle difference of these proteins. | ''Figure 4: Alignement of myosin VI structure (2BKI) (<span style="color:green">'''in green'''</span>) with myosin V (1OE9) (<span style="color:blue">'''blue'''</span>), focusing on the last helix to show the angle difference of these proteins. | ||
| - | |||
| - | Also, there is <scene name='97/973101/Insert_2_full_salt_bridges/1'>'''salt-bridge interactions'''</scene> between the converter and both lobes of CaM, with the aminoacids: | ||
| - | |||
| - | [[Image:Salt brigdes.jpg|450px]] | ||
| - | |||
| - | |||
| - | |||
| - | Figure adapted from Ménétrey ''et al.'', 2005. | ||
== 3D structure == | == 3D structure == | ||
Revision as of 14:29, 14 July 2023
Myosin VI nucleotide-free (MDinsert2-IQ) crystal structure
| |||||||||||