We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
Green Fluorescent Protein
From Proteopedia
(Difference between revisions)
| Line 26: | Line 26: | ||
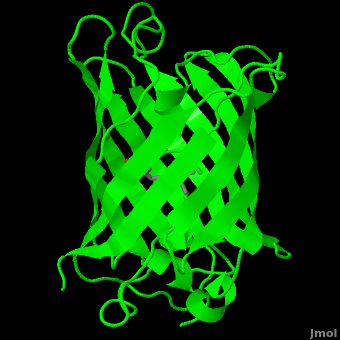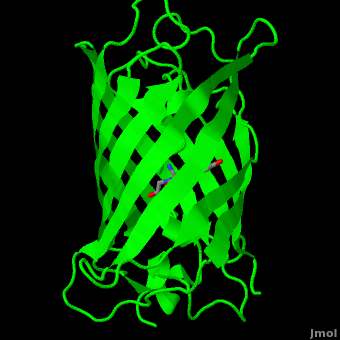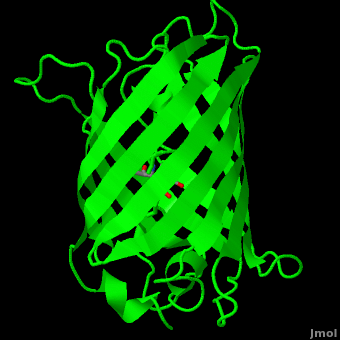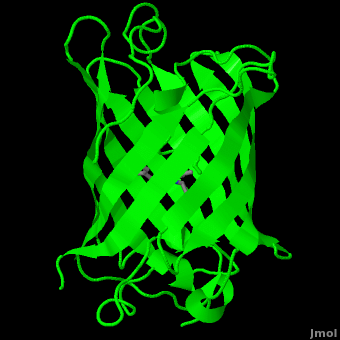
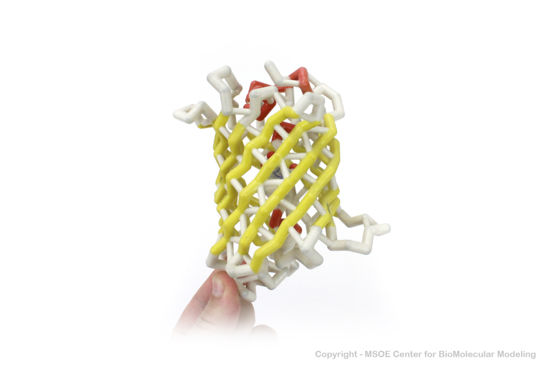
<scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2'>secondary structure</scene> of five α-helices and one eleven-stranded β-pleated sheet,<ref name="PDBsum" /> where each strand contains nine to thirteen residues each.<ref name="Ormo" /> (To view the primary and secondary structure of GFP, go to https://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=1EMA.) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.<ref name="Phillips">PMID: 9434902</ref> This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.<ref name="Yang" /> In GFP, the structure is so regular that <scene name='Green_Fluorescent_Protein/Water_stripes/1'>"stripes"</scene> of water molecules (red) can be seen following the structure of the barrel.<ref name="Phillips" /> Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,<ref name="Ormo" /> creating what is referred to as a “β-can” formation.<ref name="Phillips" /> The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.<ref name="Phillips" /> Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.<ref name="Yang" /> | <scene name='Green_Fluorescent_Protein/1ema_gfp_barrel/2'>secondary structure</scene> of five α-helices and one eleven-stranded β-pleated sheet,<ref name="PDBsum" /> where each strand contains nine to thirteen residues each.<ref name="Ormo" /> (To view the primary and secondary structure of GFP, go to https://www.rcsb.org/pdb/explore/remediatedSequence.do?structureId=1EMA.) These β-strands display an almost “seamless symmetry” in which only two of the strands vary in structural content.<ref name="Phillips">PMID: 9434902</ref> This β-sheet conforms itself through regular hydrogen bonding into a β-barrel.<ref name="Yang" /> In GFP, the structure is so regular that <scene name='Green_Fluorescent_Protein/Water_stripes/1'>"stripes"</scene> of water molecules (red) can be seen following the structure of the barrel.<ref name="Phillips" /> Together with the α-helices at either end of the molecule, a nearly perfect cylinder is produced, 42Å long and 24Å in diameter,<ref name="Ormo" /> creating what is referred to as a “β-can” formation.<ref name="Phillips" /> The short helical segments at either end of the cylinder form “caps” to further protect the interior of the β-barrel.<ref name="Phillips" /> Overall stability is maintained by this β-can structure, helping to resist unfolding from heat and other denaturants.<ref name="Yang" /> | ||
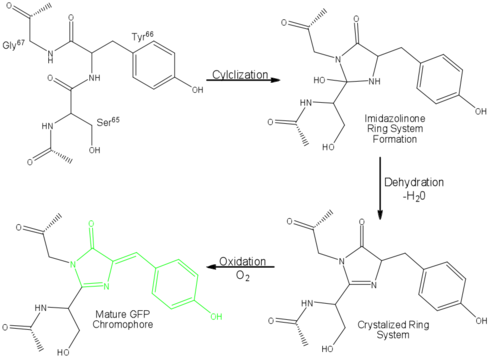
| - | One < | + | One <jmol><jmolLink><script>script "/scripts/Green_Fluorescent_Protein/Central_helix/1.spt"; ppdiaCaptionCmd = "changeCaption('The central helix (shown in red) contains the fluorophore and runs through the barrel (shown as white transparent strands) along its axis (PDB-ID [[1ema]]). ','white','black');";javascript @ppdiaCaptionCmd;model 2;</script><text>α-helix</text></jmolLink></jmol> can be found running through the central axis of the β-barrel,<ref name="Haldar" /> roughly <scene name='Green_Fluorescent_Protein/Perpendicular/1'>perpendicular</scene> to the symmetry axis of the barrel.<ref name="Ormo">Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the ''Aequorea victoria'' green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.</ref> This helix is extremely important as it contains the fluorophore responsible for fluorescence.<ref name="Yang" /><ref name="Haldar" /> |
The fluorophore is part of the polypeptide chain (i.e. covalently connencted). If you press the buttom below, it will show the connection. | The fluorophore is part of the polypeptide chain (i.e. covalently connencted). If you press the buttom below, it will show the connection. | ||
Revision as of 16:49, 5 August 2023
| |||||||||||
Reference for this Structure
Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
References
- ↑ 1.0 1.1 [1], Protein Database (PDBsum): 1ema. European Bioinformatics (EBI); 2009.
- ↑ 2.00 2.01 2.02 2.03 2.04 2.05 2.06 2.07 2.08 2.09 2.10 2.11 2.12 2.13 2.14 2.15 2.16 2.17 [2], Yang F, Moss LG, Phillips GN Jr. 1996. The molecular structure of green fluorescent protein. Biotechnology. 14: 1246-1251. DOI 10.1038/nbt1096-1246.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 Tsien, Roger Y. 1998. The Green Fluorescent Protein. Annual Review in Biochemistry. 67:509-544.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 4.6 4.7 [3], Haldar S, Chattopadhyay A. 2009. The green journey. J Fluoresc. 19:1-2. DOI 10.1007/s10895-008-0455-6; biographical background on Douglas Prasher, Martin Chalfie and Roger Tsien.
- ↑ 5.0 5.1 5.2 5.3 [4], Shimomura O. The discovery of green fluorescent protein. Nobel Prize Lecture; 2009;; biographical background at Wikipedia.
- ↑ [5],Cowles D, Cowles J. Aequorea victoria. 2007. Walla Wall University.
- ↑ Primary structure at www.ebi.aci.uk.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273(5280):1392-1395. DOI 10.1126/science.273.5280.1392.
- ↑ 9.00 9.01 9.02 9.03 9.04 9.05 9.06 9.07 9.08 9.09 9.10 9.11 9.12 9.13 Phillips GN Jr. Structure and dynamics of green fluorescent protein. Curr Opin Struct Biol. 1997 Dec;7(6):821-7. PMID:9434902
- ↑ Andrews BT, Gosavi S, Finke JM, Onuchic JN, Jennings PA. The dual-basin landscape in GFP folding. Proc Natl Acad Sci U S A. 2008 Aug 26;105(34):12283-8. Epub 2008 Aug 19. PMID:18713871
- ↑ 11.0 11.1 11.2 11.3 11.4 11.5 [6],Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien R. 1995. Understanding, improving, and using green fluorescent protein. Trends in Biochemical Sciences. 20(11): 448-455. DOI 0.1016/S0968-0004(00)89099-4.
- ↑ Bublitz G, King BA, Boxer SG. 1998. Electronic structure of the chromophore in green fluorescent protein (GFP). Journal of the American Chemical Society. 120(36): 9370-9371. DOI 10.1021/ja98160e.
- ↑ van Thor JJ, Sage, JT. 2006. Charge transfer in green fluorescent protein. Photochemical & Photobiological Sciences. 5:597-602. DOI 10.1039/b516525c.
- ↑ 14.0 14.1 Lammich L, Petersen MA, Nielsen MB, Andersen LH. The gas-phase absorption spectrum of a neutral GFP model chromophore. Biophys J. 2007 Jan 1;92(1):201-7. Epub 2006 Oct 13. PMID:17040991 doi:10.1529/biophysj.106.093674
- ↑ Information about edge-face (CH/π) interactions.
- ↑ Fang C, Frontiera RR, Tran R, Mathies RA. Mapping GFP structure evolution during proton transfer with femtosecond Raman spectroscopy. Nature. 2009 Nov 12;462(7270):200-4. PMID:19907490 doi:10.1038/nature08527
Additional Resources
- For additional information, see: Colored & Bioluminescent Proteins
- First Glance
- PDBsum: 1ema
- RCSB PDB 1ema
- OCA
- UniProt: P42212
- Scop: P42212
- CATH: 1emaA00
- Pfam: PF01353
- InterPro: IPR000786
- GFP featured at the Molecule of the Month series of tutorials by David Goodsell.
- Inside green fluorescent protein - editor's summary that accompanied structural detail of GFP chromophore on the cover of Nature.
Proteopedia Page Contributors and Editors (what is this?)
Michal Harel, Wayne Decatur, Karsten Theis, Eran Hodis, Laura Carbone, Karl Oberholser, Mark Hoelzer, Joel L. Sussman, Alexander Berchansky, Jaime Prilusky, Marius Mihasan, Joseph M. Steinberger, David Canner