|
|
| Line 3: |
Line 3: |
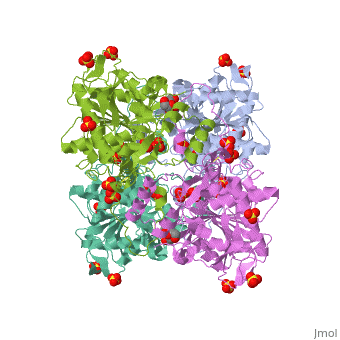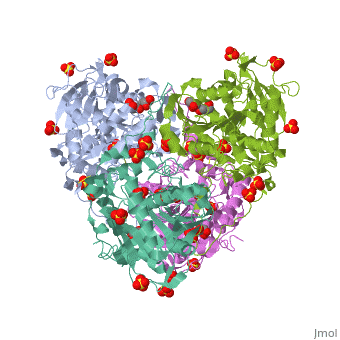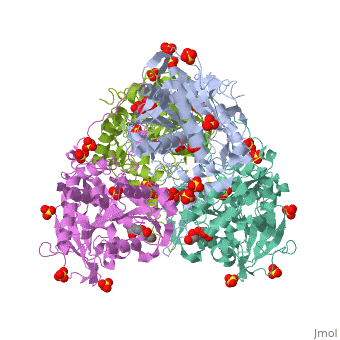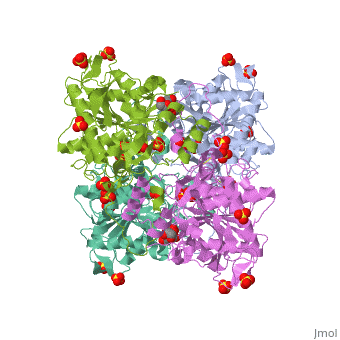
| | <StructureSection load='5hz2' size='340' side='right'caption='[[5hz2]], [[Resolution|resolution]] 1.80Å' scene=''> | | <StructureSection load='5hz2' size='340' side='right'caption='[[5hz2]], [[Resolution|resolution]] 1.80Å' scene=''> |
| | == Structural highlights == | | == Structural highlights == |
| - | <table><tr><td colspan='2'>[[5hz2]] is a 1 chain structure with sequence from [http://en.wikipedia.org/wiki/Alcaligenes_eutropha_h16 Alcaligenes eutropha h16]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5HZ2 OCA]. For a <b>guided tour on the structure components</b> use [http://proteopedia.org/fgij/fg.htm?mol=5HZ2 FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[5hz2]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Cupriavidus_necator_H16 Cupriavidus necator H16]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=5HZ2 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=5HZ2 FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.8Å</td></tr> |
| - | <tr id='gene'><td class="sblockLbl"><b>[[Gene|Gene:]]</b></td><td class="sblockDat">phbC, H16_A1437 ([http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Info&srchmode=5&id=381666 Alcaligenes eutropha H16])</td></tr> | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=GOL:GLYCEROL'>GOL</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://proteopedia.org/fgij/fg.htm?mol=5hz2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5hz2 OCA], [http://pdbe.org/5hz2 PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=5hz2 RCSB], [http://www.ebi.ac.uk/pdbsum/5hz2 PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=5hz2 ProSAT]</span></td></tr> | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=5hz2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=5hz2 OCA], [https://pdbe.org/5hz2 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=5hz2 RCSB], [https://www.ebi.ac.uk/pdbsum/5hz2 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=5hz2 ProSAT]</span></td></tr> |
| | </table> | | </table> |
| | == Function == | | == Function == |
| - | [[http://www.uniprot.org/uniprot/PHBC_CUPNH PHBC_CUPNH]] Polymerizes (R)-3-hydroxybutyryl-CoA to create polyhydroxybutyrate (PHB) which consists of thousands of hydroxybutyrate molecules linked end to end. PHB serves as an intracellular energy reserve material when cells grow under conditions of nutrient limitation.<ref>PMID:2670936</ref> | + | [https://www.uniprot.org/uniprot/PHAC_CUPNH PHAC_CUPNH] Polymerizes (R)-3-hydroxybutyryl-CoA to create polyhydroxybutyrate (PHB) which consists of thousands of hydroxybutyrate molecules linked end to end. PHB serves as an intracellular energy reserve material when cells grow under conditions of nutrient limitation.<ref>PMID:2670936</ref> |
| | <div style="background-color:#fffaf0;"> | | <div style="background-color:#fffaf0;"> |
| | == Publication Abstract from PubMed == | | == Publication Abstract from PubMed == |
| Line 26: |
Line 26: |
| | __TOC__ | | __TOC__ |
| | </StructureSection> | | </StructureSection> |
| - | [[Category: Alcaligenes eutropha h16]] | + | [[Category: Cupriavidus necator H16]] |
| | [[Category: Large Structures]] | | [[Category: Large Structures]] |
| - | [[Category: Kim, J]] | + | [[Category: Kim J]] |
| - | [[Category: Kim, K J]] | + | [[Category: Kim K-J]] |
| - | [[Category: Transferase]]
| + | |
| Structural highlights
Function
PHAC_CUPNH Polymerizes (R)-3-hydroxybutyryl-CoA to create polyhydroxybutyrate (PHB) which consists of thousands of hydroxybutyrate molecules linked end to end. PHB serves as an intracellular energy reserve material when cells grow under conditions of nutrient limitation.[1]
Publication Abstract from PubMed
Polyhydroxyalkanoates (PHAs) are natural polyesters synthesized by numerous microorganisms as energy and reducing power storage materials, and have attracted much attention as substitutes for petroleum-based plastics. Here, we report the first crystal structure of Ralstonia eutropha PHA synthase at 1.8 A resolution and structure-based mechanisms for PHA polymerization. RePhaC1 contains two distinct domains, the N-terminal (RePhaC1ND ) and C-terminal domains (RePhaC1CD ), and exists as a dimer. RePhaC1CD catalyzes polymerization via non-processive ping-pong mechanism using a Cys-His-Asp catalytic triad. Molecular docking simulation of 3-hydroxybutyryl-CoA to the active site of RePhaC1CD reveals residues involved in the formation of 3-hydroxybutyryl-CoA binding pocket and substrate binding tunnel. Comparative analysis with other polymerases elucidates how different classes of PHA synthases show different substrate specificities. Furthermore, we attempted structure-based protein engineering and developed a RePhaC1 mutant with enhanced PHA synthase activity.
Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms.,Kim J, Kim YJ, Choi SY, Lee SY, Kim KJ Biotechnol J. 2016 Nov 3. doi: 10.1002/biot.201600648. PMID:27808482[2]
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
See Also
References
- ↑ Peoples OP, Sinskey AJ. Poly-beta-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J Biol Chem. 1989 Sep 15;264(26):15298-303. PMID:2670936
- ↑ Kim J, Kim YJ, Choi SY, Lee SY, Kim KJ. Crystal structure of Ralstonia eutropha polyhydroxyalkanoate synthase C-terminal domain and reaction mechanisms. Biotechnol J. 2016 Nov 3. doi: 10.1002/biot.201600648. PMID:27808482 doi:http://dx.doi.org/10.1002/biot.201600648
|