This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Phospholipase D
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
</StructureSection> | </StructureSection> | ||
| - | ==3D structures of phospholipase D== | ||
| - | |||
| - | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| - | |||
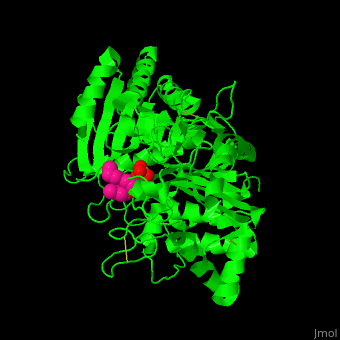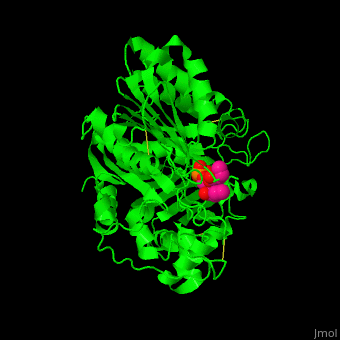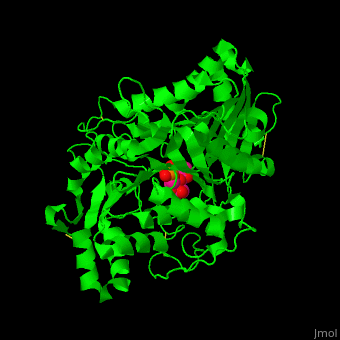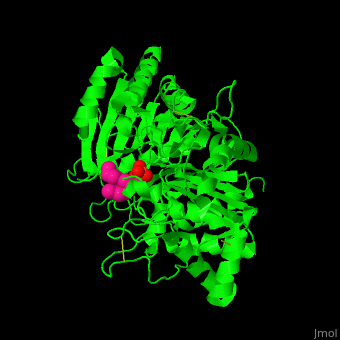
| - | [[6u8z]] – hPLD1 catalytic domain 330-500, 643-1074 – human<br /> | ||
| - | [[6ohr]] – hPLD1 catalytic domain + inhibitor<br /> | ||
| - | [[6ohm]], [[6oho]] – hPLD2 catalytic domain 294-933<br /> | ||
| - | [[6ohq]], [[6ohs]], [[6ohp]] – hPLD2 catalytic domain + inhibitor<br /> | ||
| - | [[4rw3]] – bsPLD – brown spider<br /> | ||
| - | [[4rw5]] – bsPLD (mutant)<br /> | ||
| - | [[3rlg]], [[3rlh]] – PLD (mutant) – ''Loxosceles intemedia''<br /> | ||
| - | [[4q6x]] – PLD – cave spider<br /> | ||
| - | [[2ze4]], [[1v0r]], [[1v0s]], [[1f0i]] – SaPLD – ''Streptomyces antibioticus''<br /> | ||
| - | [[7jrb]] – SaPLD (mutant) <br /> | ||
| - | [[1v0v]], [[1v0w]] – SaPLD + phosphite<br /> | ||
| - | [[1v0y]] – SaPLD + phosphatidylcholine derivative<br /> | ||
| - | [[2ze9]] – SaPLD (mutant) + phosphatidylcholine<br /> | ||
| - | [[1v0t]], [[1v0u]] – SaPLD + glycerophosphate<br /> | ||
| - | [[7jrc]] – SaPLD (mutant) + phosphate<br /> | ||
| - | [[7js5]], [[7js7]] – SaPLD (mutant) + inositol phosphate<br /> | ||
| - | [[7jru]], [[7jrv]], [[7jrw]] – SaPLD (mutant) + phosphatidic acid<br /> | ||
| - | [[6kz9]] – AtPLD a 1 – ''Arabidopsis thaliana''<br /> | ||
| - | [[6kz8]] – AtPLD a 1 + phosphatidic acid <br /> | ||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | [[Category:Topic Page]] | ||
Current revision
| |||||||||||
References
- ↑ McDermott M, Wakelam MJ, Morris AJ. Phospholipase D. Biochem Cell Biol. 2004 Feb;82(1):225-53. PMID:15052340 doi:http://dx.doi.org/10.1139/o03-079
- ↑ Oliveira TG, Di Paolo G. Phospholipase D in brain function and Alzheimer's disease. Biochim Biophys Acta. 2010 Aug;1801(8):799-805. doi:, 10.1016/j.bbalip.2010.04.004. Epub 2010 Apr 23. PMID:20399893 doi:http://dx.doi.org/10.1016/j.bbalip.2010.04.004
- ↑ Leiros I, McSweeney S, Hough E. The reaction mechanism of phospholipase D from Streptomyces sp. strain PMF. Snapshots along the reaction pathway reveal a pentacoordinate reaction intermediate and an unexpected final product. J Mol Biol. 2004 Jun 11;339(4):805-20. PMID:15165852 doi:http://dx.doi.org/10.1016/j.jmb.2004.04.003