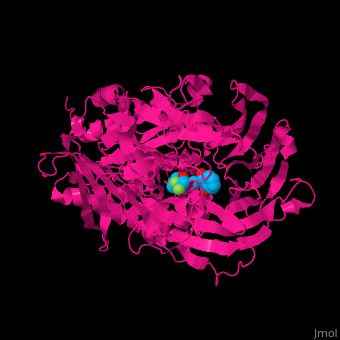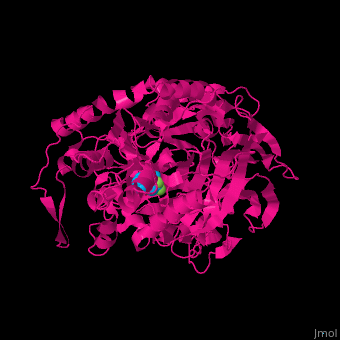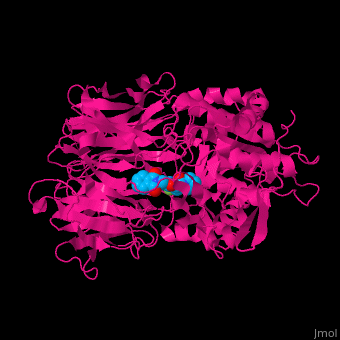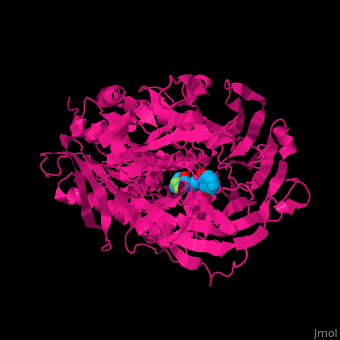
Prolyl endopeptidases (PEPs) are a class of serine proteases which cleave peptide bonds on the C-terminal side of internal proline residues.[1]
Structure
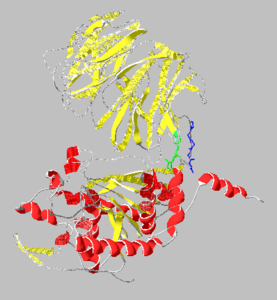
Catalytic(bottom) and β-propeller domains(top) with peptide linkages between the domains shown in green and blue
Prolyl endopeptidases are relatively large enzymes(75 kDa) and contain two distinct domains: a catalytic domain and a β-propeller domain[2].
β-Propeller Domain
The β-propeller domain is made up of repeated antiparallel β-sheets with connecting peptide strands which form a tight lid over the active site located on the catalytic domain. The β-propeller domain is cylindrical in structure and contains a very tight central channel through its core that is roughly 4Å in diameter in the resting state. It has been found that this domain is roughly 2% conserved indicating that it likely doesnt play a major role in substrate binding.[2]
Catalytic Domain
The catalytic domain mainly consists of an α/β hydrolase fold which is a series of 8 strands connected by helices in a fold common to many enzymes. The catalytic domain was found to be much more conserved than the β-propeller domain(~50%).[3] The catalytic site contains a catalytic triad of Ser-Asp-His and lies in a cavity near the interface to the β-propeller domain.[1]
Binding Mechanism
From observed interactions between the β-propeller and catalytic domains several possible substrate binding mechanism have been proposed.
The initial theory of substrate binding was that the substrate would be able to travel through the central channel in the β-propeller domain to the active site. This theory was proved less likely as the central channel is only 4Å wide in its relaxed state compared to the medium length peptides catalyzed by PEP which are 6-12Å in diameter.[4] This theory is still possible as a conformational change could allow the much larger substrate to travel through the β-propeller domain.
Another proposed mechanism of substrate binding is that the β-propeller domain acts as a gate to the active site and that the whole domain moves during a conformational change. This theory has gathered support as a structure for the PEP of Sphingomonas capsulata clearly shows the β-propeller in an open configuration connected to the catalytic domain by two peptide strands on the same side of the enzyme forming a hinge.[3]
Both mechanistic binding theories account for the observed activity of PEP as being limited to acting on small peptides of less than 30 amino acids.
Inhibition
Initial classification of prolyl endopeptidases as serine proteases was due to their inhibition by DFP.[2]
Structural data for Myxococcus xanthus determined by [1] shows that a bound inhibitor causes the β-propeller domain to become tightly associated with the catalytic domain and subsequently block the active site to inhibit catalysis.
Function
Although the physiological function of PEPs are not entirely understood there are several proposed functions based on their activity and localization.
PEPs are thought to have a role in the degradation of neuropeptides due to the high concentration of PEPs in the brain and the fact that PEPs have been shown to degrade several neuropeptides(vasopressin, β-endorphin, thyroliberin). The distribution of PEP in the brain has been found to be similar to that of certain receptors of neuropeptides which supports PEPs being involved in the degradation of neuropeptide transmitters.[3]
PEPs may also have a more general role in the degradation of peptides as PEP activity is found in most major organs and many other peptidases cannot cleave proline residues.
Pharmaceutical Possibilities
Both human and microbial PEP have been the focus of research into their viability as therapeutic agents for several diseases. The ability of PEPs to cleave internal proline peptide bonds is of interest in the treatment of Celiac Disease which is caused by a reaction to gluten, a proline rich protein found in wheat and wheat subspecies.[2] PEPs have also been linked to several neurological disorders due to high activity in the brain and a proposed role in the degradation of neuropeptides.[3]
Celiac Disease
Celiac Disease(CD) is a genetic disorder marked by diarrhea, fatigue, weight loss, and villous atrophy due to the consumption of certain proteins such as gluten and other prolamins found in wheat and wheat subspecies. The symptoms of CD are due to an auto-immune inflammatory reaction that occurs in the small intestine due to prolamins that are rich in proline residues and known to be resistant to many proteases.
There is currently no cure for Celiac Disease and the main treatment is to simply avoid eating foods containing prolamins.
Prolyl endopeptidases are a promising therapeutic agent due to their ability to degrade proline rich prolamins and inhibit the inflammatory reaction. An early idea for treatment is to orally administer certain microbial PEPs which could directly degrade prolamins in the intestine and reduce the auto-immune response. The key for this therapy would be to to protect the PEPs from being degraded by gastric acids while allowing them to be able to be functional in the small intestine once the acidity is decreased.[5]
Research by Ehren, Govindarajan, Morón, Minshull, and Khosla has shown the ability to engineer the prolyl endopeptidase of Sphingomonas capsulata to increase the activity of this PEP under simulated gastric conditions showing the relevance of PEPs as a potential therapeutic agent for Celiac Disease.[6]
Neurological Disorders
As well as having a proposed role in the degradation of neuropeptides PEPs have also been linked to several specific neurological disorders. It has been found that patients suffering from depression had decreased PEP activity and those suffering from psychosis have increased PEP activity showing the importance of maintaining a balanced level of PEP activity in the brain. PEP inhibitors are a current point of research in hopes of finding ways to balance brain PEP activity.[3]
Structural highlights
The [7]. Triad is colored in salmon.
3D structures of prolyl endopeptidase
Updated on 13-September-2023
References
- ↑ 1.0 1.1 1.2 Shan L, Mathews II, Khosla C. Structural and mechanistic analysis of two prolyl endopeptidases: role of interdomain dynamics in catalysis and specificity. Proc Natl Acad Sci U S A. 2005 Mar 8;102(10):3599-604. Epub 2005 Feb 28. PMID:15738423
- ↑ 2.0 2.1 2.2 2.3 Besedin DV, Rudenskaia GN. [Proline-specific endopeptidases] Bioorg Khim. 2003 Jan-Feb;29(1):3-20. PMID:12658988
- ↑ 3.0 3.1 3.2 3.3 3.4 Gass J, Khosla C. Prolyl endopeptidases. Cell Mol Life Sci. 2007 Feb;64(3):345-55. PMID:17160352 doi:10.1007/s00018-006-6317-y
- ↑ Shan L, Marti T, Sollid LM, Gray GM, Khosla C. Comparative biochemical analysis of three bacterial prolyl endopeptidases: implications for coeliac sprue. Biochem J. 2004 Oct 15;383(Pt 2):311-8. PMID:15245330 doi:10.1042/BJ20040907
- ↑ Ehren J, Moron B, Martin E, Bethune MT, Gray GM, Khosla C. A food-grade enzyme preparation with modest gluten detoxification properties. PLoS One. 2009 Jul 21;4(7):e6313. PMID:19621078 doi:10.1371/journal.pone.0006313
- ↑ Ehren J, Govindarajan S, Moron B, Minshull J, Khosla C. Protein engineering of improved prolyl endopeptidases for celiac sprue therapy. Protein Eng Des Sel. 2008 Dec;21(12):699-707. Epub 2008 Oct 4. PMID:18836204 doi:10.1093/protein/gzn050
- ↑ Kanai K, Aranyi P, Bocskei Z, Ferenczy G, Harmat V, Simon K, Batori S, Naray-Szabo G, Hermecz I. Prolyl oligopeptidase inhibition by N-acyl-pro-pyrrolidine-type molecules. J Med Chem. 2008 Dec 11;51(23):7514-22. PMID:19006380 doi:10.1021/jm800944x