We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
2vj1
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
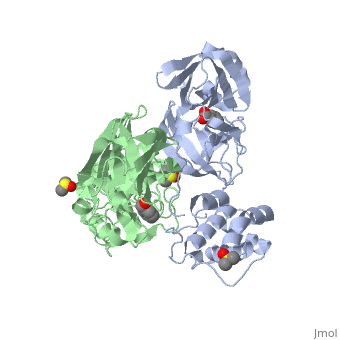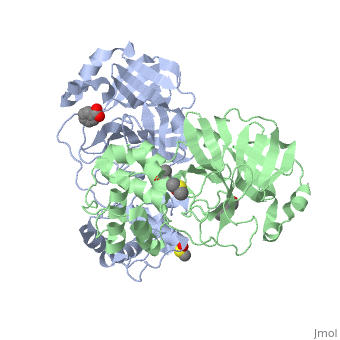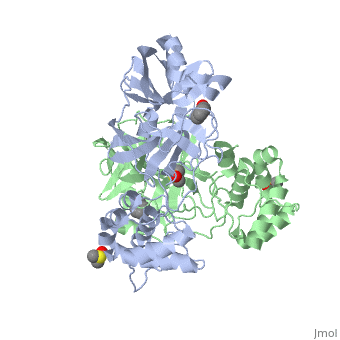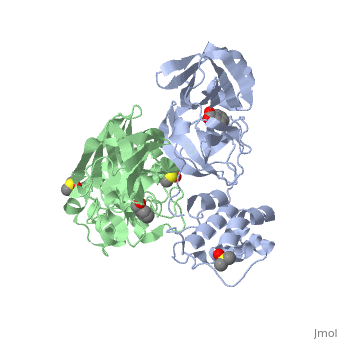
<StructureSection load='2vj1' size='340' side='right'caption='[[2vj1]], [[Resolution|resolution]] 2.25Å' scene=''> | <StructureSection load='2vj1' size='340' side='right'caption='[[2vj1]], [[Resolution|resolution]] 2.25Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[2vj1]] is a 2 chain structure. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=2cgg 2cgg]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2VJ1 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2VJ1 FirstGlance]. <br> | + | <table><tr><td colspan='2'>[[2vj1]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Severe_acute_respiratory_syndrome-related_coronavirus Severe acute respiratory syndrome-related coronavirus]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=2cgg 2cgg]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2VJ1 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2VJ1 FirstGlance]. <br> |
| - | </td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BEZ:BENZOIC+ACID'>BEZ</scene>, <scene name='pdbligand=DMS:DIMETHYL+SULFOXIDE'>DMS</scene>, <scene name='pdbligand=XP1:4-(DIMETHYLAMINO)BENZOIC+ACID'>XP1</scene></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.25Å</td></tr> |
| + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BEZ:BENZOIC+ACID'>BEZ</scene>, <scene name='pdbligand=DMS:DIMETHYL+SULFOXIDE'>DMS</scene>, <scene name='pdbligand=XP1:4-(DIMETHYLAMINO)BENZOIC+ACID'>XP1</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2vj1 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2vj1 OCA], [https://pdbe.org/2vj1 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2vj1 RCSB], [https://www.ebi.ac.uk/pdbsum/2vj1 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2vj1 ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2vj1 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2vj1 OCA], [https://pdbe.org/2vj1 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2vj1 RCSB], [https://www.ebi.ac.uk/pdbsum/2vj1 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2vj1 ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/R1AB_SARS R1AB_SARS] Multifunctional protein involved in the transcription and replication of viral RNAs. Contains the proteinases responsible for the cleavages of the polyprotein. Inhibits host translation by interacting with the 40S ribosomal subunit. The nsp1-40S ribosome complex further induces an endonucleolytic cleavage near the 5'UTR of host mRNAs, targeting them for degradation. Viral mRNAs are not susceptible to nsp1-mediated endonucleolytic RNA cleavage thanks to the presence of a 5'-end leader sequence and are therefore protected from degradation. By suppressing host gene expression, nsp1 facilitates efficient viral gene expression in infected cells and evasion from host immune response (PubMed:23035226). May disrupt nuclear pore function by binding and displacing host NUP93 (PubMed:30943371).<ref>PMID:23035226</ref> <ref>PMID:30943371</ref> May play a role in the modulation of host cell survival signaling pathway by interacting with host PHB and PHB2. Indeed, these two proteins play a role in maintaining the functional integrity of the mitochondria and protecting cells from various stresses.<ref>PMID:19640993</ref> Responsible for the cleavages located at the N-terminus of the replicase polyprotein. In addition, PL-PRO possesses a deubiquitinating/deISGylating activity and processes both 'Lys-48'- and 'Lys-63'-linked polyubiquitin chains from cellular substrates (PubMed:17692280). Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication. Nsp3, nsp4 and nsp6 together are sufficient to form DMV (PubMed:24410069). Antagonizes innate immune induction of type I interferon by blocking the phosphorylation, dimerization and subsequent nuclear translocation of host IRF3 (PubMed:19369340, PubMed:24622840). Prevents also host NF-kappa-B signaling.<ref>PMID:16271890</ref> <ref>PMID:17692280</ref> <ref>PMID:19369340</ref> <ref>PMID:24622840</ref> <ref>PMID:24410069</ref> Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication. Alone appears incapable to induce membrane curvature, but together with nsp3 is able to induce paired membranes. Nsp3, nsp4 and nsp6 together are sufficient to form DMV.<ref>PMID:23943763</ref> <ref>PMID:24410069</ref> Cleaves the C-terminus of replicase polyprotein at 11 sites. Recognizes substrates containing the core sequence [ILMVF]-Q-|-[SGACN]. Also able to bind an ADP-ribose-1''-phosphate (ADRP). May cleave host ATP6V1G1 thereby modifying host vacuoles intracellular pH.[PROSITE-ProRule:PRU00772]<ref>PMID:16226257</ref> Plays a role in host membrane rearrangement that leads to creation of cytoplasmic double-membrane vesicles (DMV) necessary for viral replication. Nsp3, nsp4 and nsp6 together are sufficient to form DMV (PubMed:24410069). Plays a role in the initial induction of autophagosomes from host reticulum endoplasmic. Later, limits the expansion of these phagosomes that are no longer able to deliver viral components to lysosomes (PubMed:24991833).<ref>PMID:24991833</ref> <ref>PMID:24410069</ref> Forms a hexadecamer with nsp8 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.<ref>PMID:22039154</ref> Forms a hexadecamer with nsp7 (8 subunits of each) that may participate in viral replication by acting as a primase. Alternatively, may synthesize substantially longer products than oligonucleotide primers.<ref>PMID:22039154</ref> May participate in viral replication by acting as a ssRNA-binding protein.<ref>PMID:19153232</ref> Plays a pivotal role in viral transcription by stimulating both nsp14 3'-5' exoribonuclease and nsp16 2'-O-methyltransferase activities. Therefore plays an essential role in viral mRNAs cap methylation.<ref>PMID:22635272</ref> Responsible for replication and transcription of the viral RNA genome.<ref>PMID:22791111</ref> Multi-functional protein with a zinc-binding domain in N-terminus displaying RNA and DNA duplex-unwinding activities with 5' to 3' polarity. Activity of helicase is dependent on magnesium.<ref>PMID:12917423</ref> <ref>PMID:22615777</ref> Enzyme possessing two different activities: an exoribonuclease activity acting on both ssRNA and dsRNA in a 3' to 5' direction and a N7-guanine methyltransferase activity (PubMed:16549795, PubMed:20421945, PubMed:22635272). Acts as a proofreading exoribonuclease for RNA replication, thereby lowering The sensitivity of the virus to RNA mutagens (PubMed:23966862, PubMed:29511076, PubMed:21593585).<ref>PMID:16549795</ref> <ref>PMID:20421945</ref> <ref>PMID:21593585</ref> <ref>PMID:22635272</ref> <ref>PMID:23966862</ref> <ref>PMID:29511076</ref> Mn(2+)-dependent, uridylate-specific enzyme, which leaves 2'-3'-cyclic phosphates 5' to the cleaved bond. Methyltransferase that mediates mRNA cap 2'-O-ribose methylation to the 5'-cap structure of viral mRNAs. N7-methyl guanosine cap is a prerequisite for binding of nsp16. Therefore plays an essential role in viral mRNAs cap methylation which is essential to evade immune system.<ref>PMID:18417574</ref> <ref>PMID:20421945</ref> | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 37: | Line 38: | ||
</StructureSection> | </StructureSection> | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | [[Category: | + | [[Category: Severe acute respiratory syndrome-related coronavirus]] |
| - | [[Category: | + | [[Category: Anemueller S]] |
| - | [[Category: | + | [[Category: Hilgenfeld R]] |
| - | [[Category: | + | [[Category: Mesters JR]] |
| - | [[Category: | + | [[Category: Pumpor K]] |
| - | [[Category: | + | [[Category: Verschueren KHG]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
A Structural View of the Inactivation of the SARS-Coronavirus Main Proteinase by Benzotriazole Esters
| |||||||||||