1ppb
From Proteopedia
| Line 1: | Line 1: | ||
[[Image:1ppb.gif|left|200px]] | [[Image:1ppb.gif|left|200px]] | ||
| - | + | <!-- | |
| - | + | The line below this paragraph, containing "STRUCTURE_1ppb", creates the "Structure Box" on the page. | |
| - | + | You may change the PDB parameter (which sets the PDB file loaded into the applet) | |
| - | + | or the SCENE parameter (which sets the initial scene displayed when the page is loaded), | |
| - | + | or leave the SCENE parameter empty for the default display. | |
| - | | | + | --> |
| - | | | + | {{STRUCTURE_1ppb| PDB=1ppb | SCENE= }} |
| - | + | ||
| - | + | ||
| - | }} | + | |
'''THE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENT''' | '''THE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENT''' | ||
| Line 27: | Line 24: | ||
[[Category: Thrombin]] | [[Category: Thrombin]] | ||
[[Category: Bode, W.]] | [[Category: Bode, W.]] | ||
| - | + | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on Sat May 3 05:20:09 2008'' | |
| - | + | ||
| - | ''Page seeded by [http://oca.weizmann.ac.il/oca OCA ] on | + | |
Revision as of 02:20, 3 May 2008
| |||||||||
| 1ppb, resolution 1.92Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| Activity: | Thrombin, with EC number 3.4.21.5 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
THE REFINED 1.9 ANGSTROMS CRYSTAL STRUCTURE OF HUMAN ALPHA-THROMBIN: INTERACTION WITH D-PHE-PRO-ARG CHLOROMETHYLKETONE AND SIGNIFICANCE OF THE TYR-PRO-PRO-TRP INSERTION SEGMENT
Overview
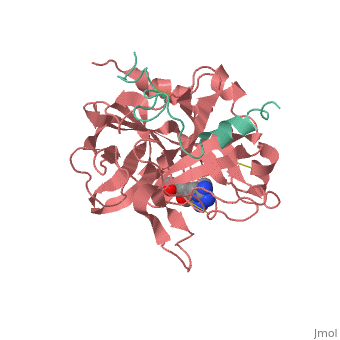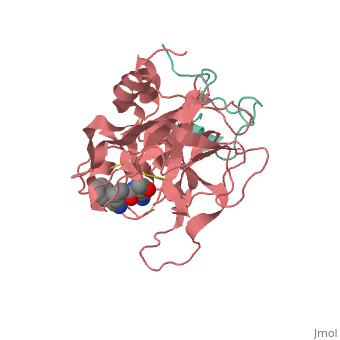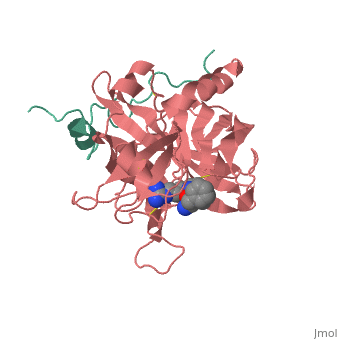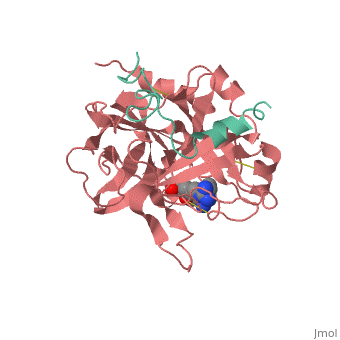
A stoichiometric complex formed between human alpha-thrombin and D-Phe-Pro-Arg chloromethylketone was crystallized in an orthorhombic crystal form. Orientation and position of a starting model derived from homologous modelling were determined by Patterson search methods. The thrombin model was completed in a cyclic modelling-crystallographic refinement procedure to a final R-value of 0.171 for X-ray data to 1.92 A. The structure is in full agreement with published cDNA sequence data. The A-chain, ordered only in its central part, is positioned along the molecular surface opposite to the active site. The B-chain exhibits the characteristic polypeptide fold of trypsin-like proteinases. Several extended insertions form, however, large protuberances; most important for interaction with macromolecular substrates is the characteristic thrombin loop around Tyr60A-Pro60B-Pro60C-Trp60D (chymotrypsinogen numbering) and the enlarged loop around the unique Trp148. The former considerably restricts the active site cleft and seems likely to be responsible for poor binding of most natural proteinase inhibitors to thrombin. The exceptional specificity of D-Phe-Pro-Arg chloromethylketone can be explained by a hydrophobic cage formed by Ile174, Trp215, Leu99, His57, Tyr60A and Trp60D. The narrow active site cleft, with a more polar base and hydrophobic rims, extends towards the arginine-rich surface of loop Lys70-Glu80 that probably represents part of the anionic binding region for hirudin and fibrinogen.
About this Structure
1PPB is a Protein complex structure of sequences from Homo sapiens. The following page contains interesting information on the relation of 1PPB with [Thrombin]. Full crystallographic information is available from OCA.
Reference
The refined 1.9 A crystal structure of human alpha-thrombin: interaction with D-Phe-Pro-Arg chloromethylketone and significance of the Tyr-Pro-Pro-Trp insertion segment., Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J, EMBO J. 1989 Nov;8(11):3467-75. PMID:2583108 Page seeded by OCA on Sat May 3 05:20:09 2008