The Initial Search
We were given a Protein with a predicted structure from Uniport and an unknown function. We are trying to find the function of the protein. We did this by first using computational tools like Blast, Dali, and Interpro to help us find potential substrates.
The first computational tool we used was Blast the results of which are shown below on Table 1. (blast figure).
From the Blast results we saw that it was from the ROK family and that it was a putative kinase.
The next tool we used was Dali shown in table 2.(Dali figure).
From the Dali results the best results we saw were from a putative kinase, glucokinase and a DNA-binding kinase. These results are what showed us that there could be potential nucleosides though we weren't for sure which one it could be until we go to docking.
The next tool we used was InterPro which can be seen in Table 3 (InterPro figure).
The InterPro figure really help us narrow down that it could be either of those to substrates because again it shown a sugar kinase and a ATPase which would mean that there is potentially a nucloeside or something to with ATP.
Molecular Docking
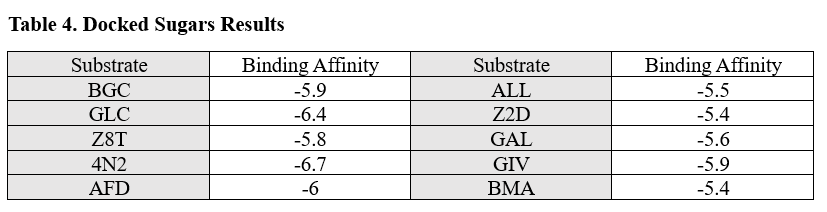
From the initial search throughout all of the computational tools, we decided that our putative kinase was potentially a glucokinase. We docked other sugars along with glucose in figure 1.
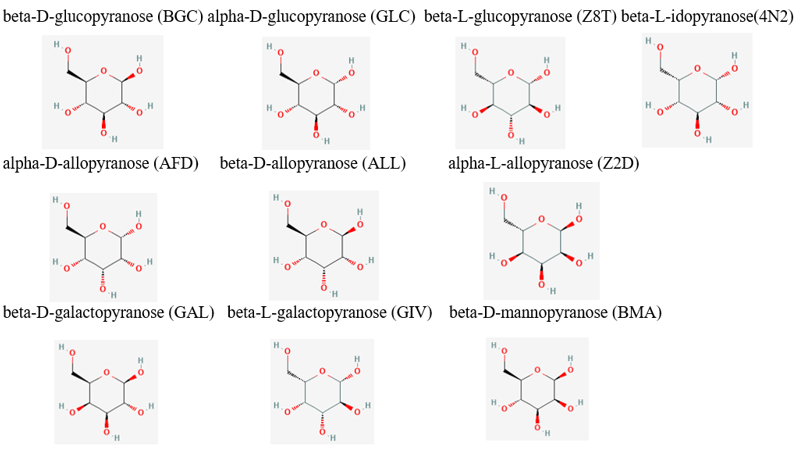
Taking in consideration of the DNA binding domain found in the InterPro results we docked DNA nitrogenous bases and nucleosides, the structures of the nitrogenous bases are shown in figure 2.
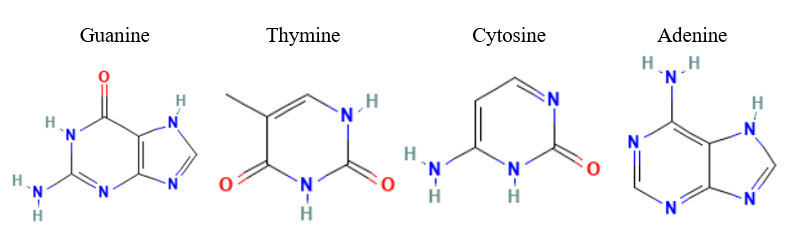
From the results of the docked sugars, shown in table 4, nitrogenous bases, and nucleosides, we determined that guanosine was a strong potential substrate but still wanted to test glucose due to the computational tools results since glucokinase was a common output in all of our searches.
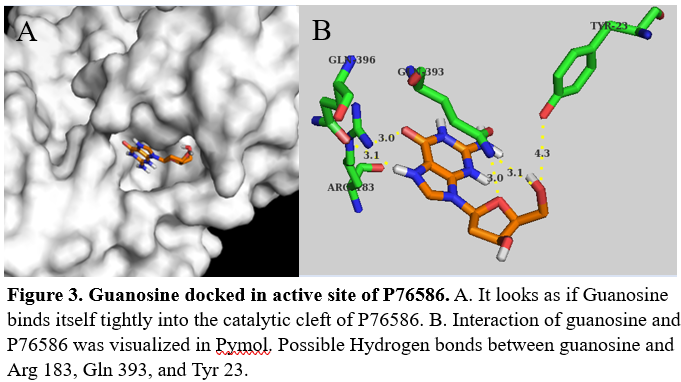
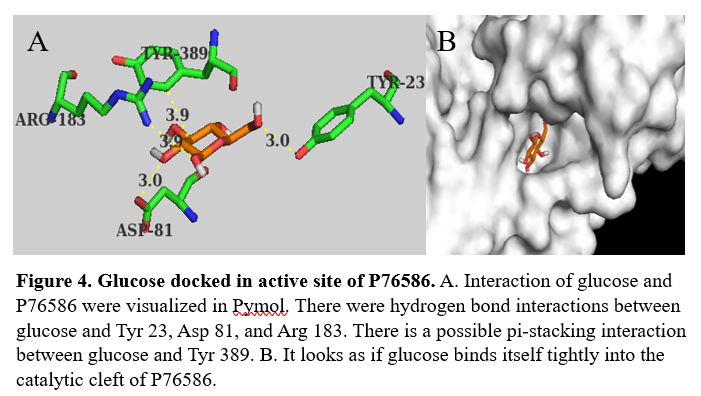
We used Pymol to visualize the intermolecular interactions in the active site with guanosine (figure 3) and glucose (figure 4).
Structural Highlights
with 397 amino acids. It's is made up of mostly alpha helices, but also has beta sheets and random coil. Through docking we were able to identify possible amino acids involved in the of P76586. Potential amino acids in the active site are Tyr23, Asp81, Arg183, Gln393, and Tyr389.
Results
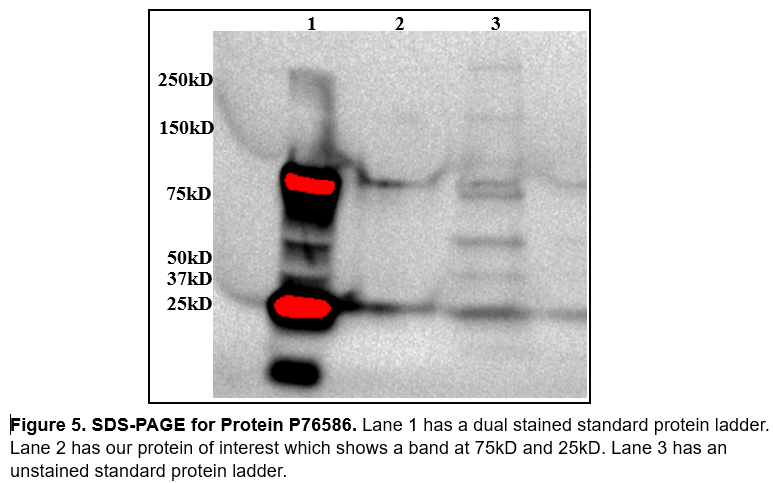
Our protein of interest has a weight of ≈44.53kD. When analyzing SDS PAGE (figure 5) we were slightly concerned we weren't working with our protein of interest. We didn't get a great image out of SDS, if there were more time we would run again with more protein in the well so that we could see it better. We also made the mistake of not including our induction samples and our fractions from protein purification using Nickle affinity chromatography. The band at 75kD and 25kD we believe are there from the imaging and is not from our actual protein sample.
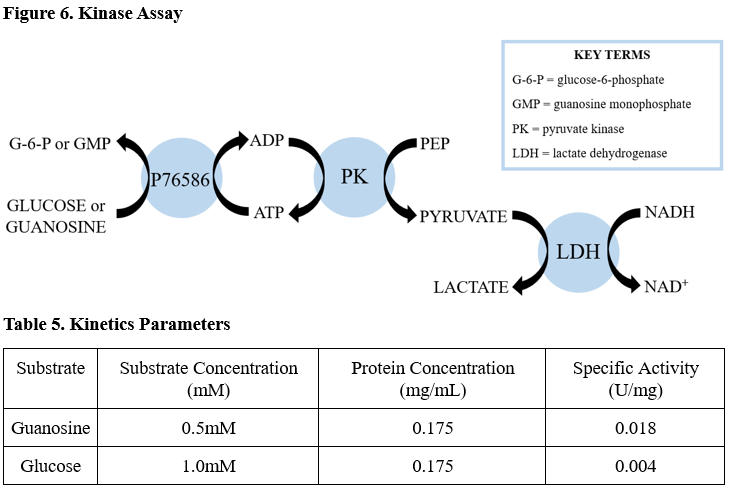
We ran two coupled kinase assays, one with glucose and one with guanosine. After calculating the specific activity of our protein with glucose and guanosine we determined there was not enough activity for glucose or guanosine to be the substrate (table 5). We didn't have to modify the kinase assay protocol for Abl Kinase other than changing the substrate and ensuring we were using a high enough substrate concentration that was greater than the protein concentration.
Conclusions
Although we determined that neither glucose or guanosine were the substrate for P76586, we are unsure if this may have been from experimental error. But looking back at the docking results I think we should have tried a wider range of substrates. Maybe a substrate that had closer hydrogen bond interactions. If we were to continue our research we would spend much more time on the docking portion in order to really dissect the active site and its interactions with the substrates. Overall this research experience was really educational and we gained a lot of skills from working in the lab and working as a team.







