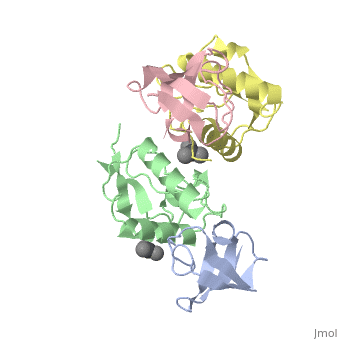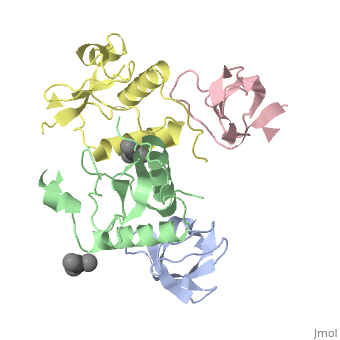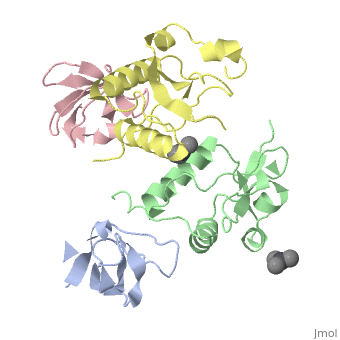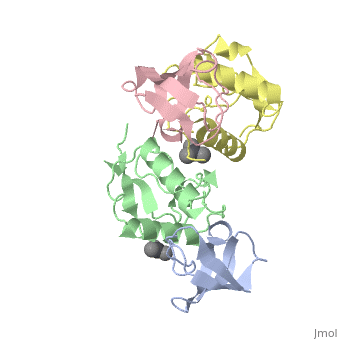We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1efn
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1efn' size='340' side='right'caption='[[1efn]], [[Resolution|resolution]] 2.50Å' scene=''> | <StructureSection load='1efn' size='340' side='right'caption='[[1efn]], [[Resolution|resolution]] 2.50Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[1efn]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1efn]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Human_immunodeficiency_virus_1 Human immunodeficiency virus 1]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1EFN OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1EFN FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=PBM:TRIMETHYL+LEAD+ION'>PBM</scene></td></tr> |
| - | + | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1efn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1efn OCA], [https://pdbe.org/1efn PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1efn RCSB], [https://www.ebi.ac.uk/pdbsum/1efn PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1efn ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1efn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1efn OCA], [https://pdbe.org/1efn PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1efn RCSB], [https://www.ebi.ac.uk/pdbsum/1efn PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1efn ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/FYN_HUMAN FYN_HUMAN] Non-receptor tyrosine-protein kinase that plays a role in many biological processes including regulation of cell growth and survival, cell adhesion, integrin-mediated signaling, cytoskeletal remodeling, cell motility, immune response and axon guidance. Inactive FYN is phosphorylated on its C-terminal tail within the catalytic domain. Following activation by PKA, the protein subsequently associates with PTK2/FAK1, allowing PTK2/FAK1 phosphorylation, activation and targeting to focal adhesions. Involved in the regulation of cell adhesion and motility through phosphorylation of CTNNB1 (beta-catenin) and CTNND1 (delta-catenin). Regulates cytoskeletal remodeling by phosphorylating several proteins including the actin regulator WAS and the microtubule-associated proteins MAP2 and MAPT. Promotes cell survival by phosphorylating AGAP2/PIKE-A and preventing its apoptotic cleavage. Participates in signal transduction pathways that regulate the integrity of the glomerular slit diaphragm (an essential part of the glomerular filter of the kidney) by phosphorylating several slit diaphragm components including NPHS1, KIRREL and TRPC6. Plays a role in neural processes by phosphorylating DPYSL2, a multifunctional adapter protein within the central nervous system, ARHGAP32, a regulator for Rho family GTPases implicated in various neural functions, and SNCA, a small pre-synaptic protein. Participates in the downstream signaling pathways that lead to T-cell differentiation and proliferation following T-cell receptor (TCR) stimulation. Also participates in negative feedback regulation of TCR signaling through phosphorylation of PAG1, thereby promoting interaction between PAG1 and CSK and recruitment of CSK to lipid rafts. CSK maintains LCK and FYN in an inactive form. Promotes CD28-induced phosphorylation of VAV1.<ref>PMID:7822789</ref> <ref>PMID:7568038</ref> <ref>PMID:11005864</ref> <ref>PMID:11162638</ref> <ref>PMID:11536198</ref> <ref>PMID:12788081</ref> <ref>PMID:12640114</ref> <ref>PMID:14761972</ref> <ref>PMID:15557120</ref> <ref>PMID:14707117</ref> <ref>PMID:15536091</ref> <ref>PMID:16387660</ref> <ref>PMID:16841086</ref> <ref>PMID:17194753</ref> <ref>PMID:18056706</ref> <ref>PMID:18258597</ref> <ref>PMID:19179337</ref> <ref>PMID:19652227</ref> <ref>PMID:20100835</ref> | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 21: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1efn ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1efn ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The crystal structure of the conserved core of HIV-1 Nef has been determined in complex with the SH3 domain of a mutant Fyn tyrosine kinase (a single amino acid substitution, Arg-96 to isoleucine), to which Nef binds tightly. The conserved PxxP sequence motif of Nef, known to be important for optimal viral replication, is part of a polyproline type II helix that engages the SH3 domain in a manner resembling closely the interaction of isolated peptides with SH3 domains. The Nef-SH3 structure also reveals how high affinity and specificity in the SH3 interaction is achieved by the presentation of the PxxP motif within the context of the folded structure of Nef. | ||
| - | |||
| - | Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain.,Lee CH, Saksela K, Mirza UA, Chait BT, Kuriyan J Cell. 1996 Jun 14;85(6):931-42. PMID:8681387<ref>PMID:8681387</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1efn" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| - | *[[Protein Nef|Protein Nef]] | + | *[[Protein Nef 3D structures|Protein Nef 3D structures]] |
*[[Tyrosine kinase 3D structures|Tyrosine kinase 3D structures]] | *[[Tyrosine kinase 3D structures|Tyrosine kinase 3D structures]] | ||
== References == | == References == | ||
| Line 38: | Line 28: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: Human]] | + | [[Category: Homo sapiens]] |
| + | [[Category: Human immunodeficiency virus 1]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | + | [[Category: Kuriyan J]] | |
| - | [[Category: Kuriyan | + | [[Category: Lee C-H]] |
| - | [[Category: Lee | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
HIV-1 NEF PROTEIN IN COMPLEX WITH R96I MUTANT FYN SH3 DOMAIN
| |||||||||||