We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1nye
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1nye' size='340' side='right'caption='[[1nye]], [[Resolution|resolution]] 2.40Å' scene=''> | <StructureSection load='1nye' size='340' side='right'caption='[[1nye]], [[Resolution|resolution]] 2.40Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
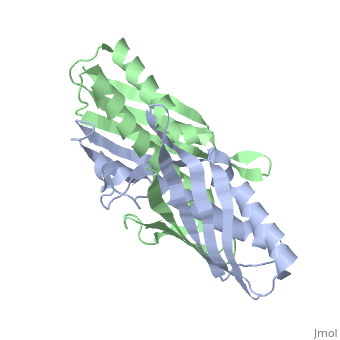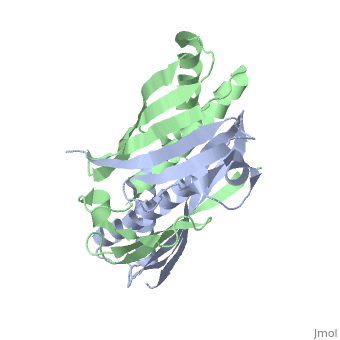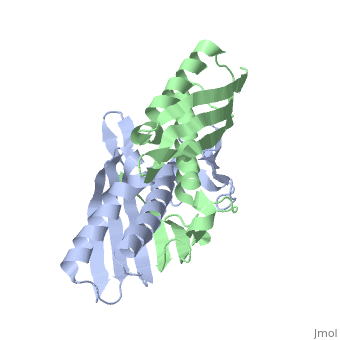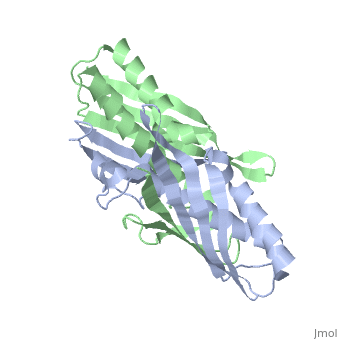
| - | <table><tr><td colspan='2'>[[1nye]] is a 6 chain structure with sequence from [ | + | <table><tr><td colspan='2'>[[1nye]] is a 6 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1NYE OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1NYE FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.4Å</td></tr> |
| - | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[ | + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1nye FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1nye OCA], [https://pdbe.org/1nye PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1nye RCSB], [https://www.ebi.ac.uk/pdbsum/1nye PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1nye ProSAT], [https://www.topsan.org/Proteins/BSGC/1nye TOPSAN]</span></td></tr> |
</table> | </table> | ||
== Function == | == Function == | ||
| - | [ | + | [https://www.uniprot.org/uniprot/OSMC_ECOLI OSMC_ECOLI] Preferentially metabolizes organic hydroperoxides over inorganic hydrogen peroxide.<ref>PMID:14627744</ref> |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 19: | Line 19: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1nye ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1nye ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The crystal structure of an osmotically inducible protein (OsmC) from Escherichia coli has been determined at 2.4 A resolution. OsmC is a representative protein of the OsmC sequence family, which is composed of three sequence subfamilies. The structure of OsmC provides a view of a salt-shock-induced protein. Two identical monomers form a cylindrically shaped dimer in which six helices are located on the inside and two six-stranded beta-sheets wrap around these helices. Structural comparison suggests that the OsmC sequence family has a peroxiredoxin function and has a unique structure compared with other peroxiredoxin families. A detailed analysis of structures and sequence comparisons in the OsmC sequence family revealed that each subfamily has unique motifs. In addition, the molecular function of the OsmC sequence family is discussed based on structural comparisons among the subfamily members. | ||
| - | |||
| - | Structure of OsmC from Escherichia coli: a salt-shock-induced protein.,Shin DH, Choi IG, Busso D, Jancarik J, Yokota H, Kim R, Kim SH Acta Crystallogr D Biol Crystallogr. 2004 May;60(Pt 5):903-11. Epub 2004, Apr 21. PMID:15103136<ref>PMID:15103136</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1nye" style="background-color:#fffaf0;"></div> | ||
== References == | == References == | ||
<references/> | <references/> | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Escherichia coli]] |
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
| - | + | [[Category: Busso D]] | |
| - | [[Category: Busso | + | [[Category: Choi I-G]] |
| - | [[Category: Choi | + | [[Category: Jancarik J]] |
| - | [[Category: Jancarik | + | [[Category: Kim R]] |
| - | [[Category: Kim | + | [[Category: Kim S-H]] |
| - | [[Category: Kim | + | [[Category: Shin DH]] |
| - | [[Category: Shin | + | [[Category: Yokota H]] |
| - | [[Category: Yokota | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Crystal structure of OsmC from E. coli
| |||||||||||
Categories: Escherichia coli | Large Structures | Busso D | Choi I-G | Jancarik J | Kim R | Kim S-H | Shin DH | Yokota H