We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1u6b
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1u6b' size='340' side='right'caption='[[1u6b]], [[Resolution|resolution]] 3.10Å' scene=''> | <StructureSection load='1u6b' size='340' side='right'caption='[[1u6b]], [[Resolution|resolution]] 3.10Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
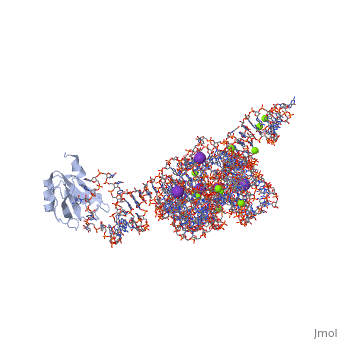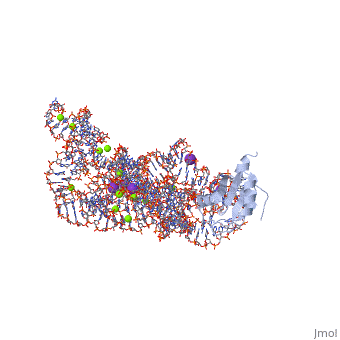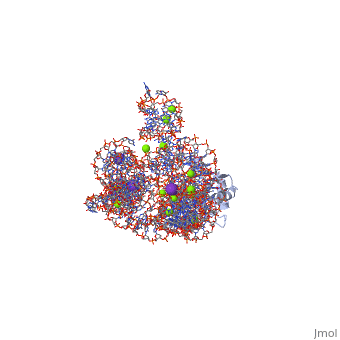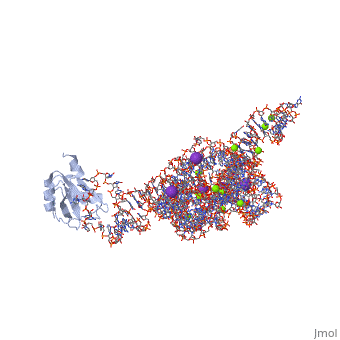
| - | <table><tr><td colspan='2'>[[1u6b]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1u6b]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. This structure supersedes the now removed PDB entry [http://oca.weizmann.ac.il/oca-bin/send-pdb?obs=1&id=1t42 1t42]. The May 2005 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''Self-splicing RNA'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_5 10.2210/rcsb_pdb/mom_2005_5]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1U6B OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1U6B FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3.1Å</td></tr> |
| - | <tr id=' | + | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=5MU:5-METHYLURIDINE+5-MONOPHOSPHATE'>5MU</scene>, <scene name='pdbligand=A23:ADENOSINE-5-PHOSPHATE-2,3-CYCLIC+PHOSPHATE'>A23</scene>, <scene name='pdbligand=GTP:GUANOSINE-5-TRIPHOSPHATE'>GTP</scene>, <scene name='pdbligand=K:POTASSIUM+ION'>K</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene></td></tr> |
| - | < | + | |
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1u6b FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1u6b OCA], [https://pdbe.org/1u6b PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1u6b RCSB], [https://www.ebi.ac.uk/pdbsum/1u6b PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1u6b ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1u6b FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1u6b OCA], [https://pdbe.org/1u6b PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1u6b RCSB], [https://www.ebi.ac.uk/pdbsum/1u6b PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1u6b ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/SNRPA_HUMAN SNRPA_HUMAN] Binds stem loop II of U1 snRNA. It is the first snRNP to interact with pre-mRNA. This interaction is required for the subsequent binding of U2 snRNP and the U4/U6/U5 tri-snRNP. In a snRNP-free form (SF-A) may be involved in coupled pre-mRNA splicing and polyadenylation process. Binds preferentially to the 5'-UGCAC-3' motif in vitro.<ref>PMID:9848648</ref> | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 21: | Line 20: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1u6b ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1u6b ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The discovery of the RNA self-splicing group I intron provided the first demonstration that not all enzymes are proteins. Here we report the X-ray crystal structure (3.1-A resolution) of a complete group I bacterial intron in complex with both the 5'- and the 3'-exons. This complex corresponds to the splicing intermediate before the exon ligation step. It reveals how the intron uses structurally unprecedented RNA motifs to select the 5'- and 3'-splice sites. The 5'-exon's 3'-OH is positioned for inline nucleophilic attack on the conformationally constrained scissile phosphate at the intron-3'-exon junction. Six phosphates from three disparate RNA strands converge to coordinate two metal ions that are asymmetrically positioned on opposing sides of the reactive phosphate. This structure represents the first splicing complex to include a complete intron, both exons and an organized active site occupied with metal ions. | ||
| - | |||
| - | Crystal structure of a self-splicing group I intron with both exons.,Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA Nature. 2004 Jul 1;430(6995):45-50. Epub 2004 Jun 2. PMID:15175762<ref>PMID:15175762</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1u6b" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
*[[Kink-turn motif|Kink-turn motif]] | *[[Kink-turn motif|Kink-turn motif]] | ||
*[[Nucleoprotein 3D structures|Nucleoprotein 3D structures]] | *[[Nucleoprotein 3D structures|Nucleoprotein 3D structures]] | ||
| - | *[[Ribozyme|Ribozyme]] | + | *[[Ribozyme 3D structures|Ribozyme 3D structures]] |
*[[User:Wayne Decatur/kink-turn motif|User:Wayne Decatur/kink-turn motif]] | *[[User:Wayne Decatur/kink-turn motif|User:Wayne Decatur/kink-turn motif]] | ||
== References == | == References == | ||
| Line 40: | Line 30: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: | + | [[Category: Homo sapiens]] |
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: Self-splicing RNA]] | [[Category: Self-splicing RNA]] | ||
| - | [[Category: Adams | + | [[Category: Adams PL]] |
| - | [[Category: Kosek | + | [[Category: Kosek AB]] |
| - | [[Category: Stahley | + | [[Category: Stahley MR]] |
| - | [[Category: Strobel | + | [[Category: Strobel SA]] |
| - | [[Category: Wang | + | [[Category: Wang J]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
CRYSTAL STRUCTURE OF A SELF-SPLICING GROUP I INTRON WITH BOTH EXONS
| |||||||||||