We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
1ytb
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
<StructureSection load='1ytb' size='340' side='right'caption='[[1ytb]], [[Resolution|resolution]] 1.80Å' scene=''> | <StructureSection load='1ytb' size='340' side='right'caption='[[1ytb]], [[Resolution|resolution]] 1.80Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
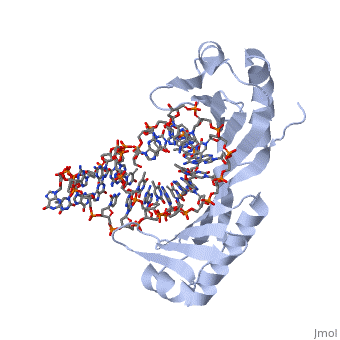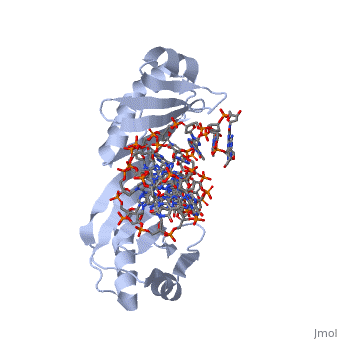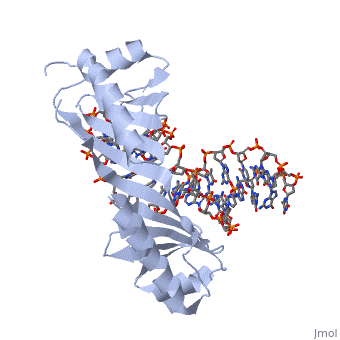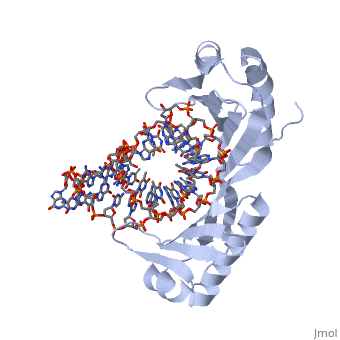
| - | <table><tr><td colspan='2'>[[1ytb]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[1ytb]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Saccharomyces_cerevisiae Saccharomyces cerevisiae]. The July 2005 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''TATA-Binding Protein'' by David S. Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2005_7 10.2210/rcsb_pdb/mom_2005_7]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1YTB OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1YTB FirstGlance]. <br> |
| - | </td></tr><tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ytb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ytb OCA], [https://pdbe.org/1ytb PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ytb RCSB], [https://www.ebi.ac.uk/pdbsum/1ytb PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ytb ProSAT]</span></td></tr> | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.8Å</td></tr> |
| + | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ytb FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ytb OCA], [https://pdbe.org/1ytb PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ytb RCSB], [https://www.ebi.ac.uk/pdbsum/1ytb PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ytb ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| - | + | [https://www.uniprot.org/uniprot/TBP_YEAST TBP_YEAST] General transcription factor that functions at the core of the DNA-binding general transcription factor complex TFIID. Binding of TFIID to a promoter (with or without TATA element) is the initial step in preinitiation complex (PIC) formation. TFIID plays a key role in the regulation of gene expression by RNA polymerase II through different activities such as transcription activator interaction, core promoter recognition and selectivity, TFIIA and TFIIB interaction, chromatin modification (histone acetylation by TAF1), facilitation of DNA opening and initiation of transcription.<ref>PMID:9618449</ref> <ref>PMID:12138208</ref> <ref>PMID:12516863</ref> | |
== Evolutionary Conservation == | == Evolutionary Conservation == | ||
[[Image:Consurf_key_small.gif|200px|right]] | [[Image:Consurf_key_small.gif|200px|right]] | ||
| Line 18: | Line 19: | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ytb ConSurf]. | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ytb ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
| - | <div style="background-color:#fffaf0;"> | ||
| - | == Publication Abstract from PubMed == | ||
| - | The 2.5 A crystal structure of a TATA-box complex with yeast TBP shows that the eight base pairs of the TATA box bind to the concave surface of TBP by bending towards the major groove with unprecedented severity. This produces a wide open, underwound, shallow minor groove which forms a primarily hydrophobic interface with the entire under-surface of the TBP saddle. The severe bend and a positive writhe radically alter the trajectory of the flanking B-form DNA. | ||
| - | |||
| - | Crystal structure of a yeast TBP/TATA-box complex.,Kim Y, Geiger JH, Hahn S, Sigler PB Nature. 1993 Oct 7;365(6446):512-20. PMID:8413604<ref>PMID:8413604</ref> | ||
| - | |||
| - | From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | ||
| - | </div> | ||
| - | <div class="pdbe-citations 1ytb" style="background-color:#fffaf0;"></div> | ||
==See Also== | ==See Also== | ||
| Line 34: | Line 26: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
| - | [[Category: Atcc 18824]] | ||
[[Category: Large Structures]] | [[Category: Large Structures]] | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
| + | [[Category: Saccharomyces cerevisiae]] | ||
[[Category: TATA-Binding Protein]] | [[Category: TATA-Binding Protein]] | ||
| - | [[Category: Geiger | + | [[Category: Geiger JH]] |
| - | [[Category: Hahn | + | [[Category: Hahn S]] |
| - | [[Category: Kim | + | [[Category: Kim Y]] |
| - | [[Category: Sigler | + | [[Category: Sigler PB]] |
| - | + | ||
| - | + | ||
Current revision
CRYSTAL STRUCTURE OF A YEAST TBP/TATA-BOX COMPLEX
| |||||||||||