This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
3ugm
From Proteopedia
(Difference between revisions)
| Line 3: | Line 3: | ||
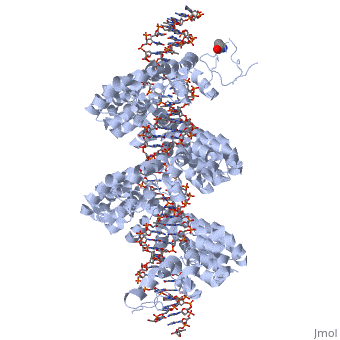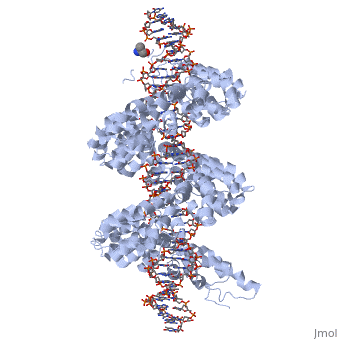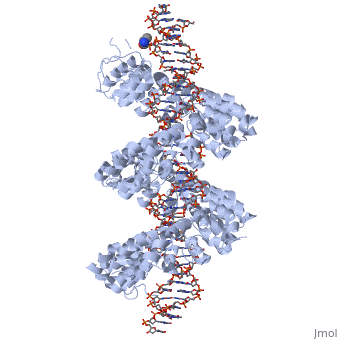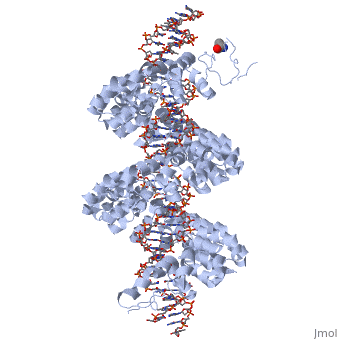
<StructureSection load='3ugm' size='340' side='right'caption='[[3ugm]], [[Resolution|resolution]] 3.00Å' scene=''> | <StructureSection load='3ugm' size='340' side='right'caption='[[3ugm]], [[Resolution|resolution]] 3.00Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
| - | <table><tr><td colspan='2'>[[3ugm]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/ | + | <table><tr><td colspan='2'>[[3ugm]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Xanthomonas_oryzae_pv._oryzae_PXO99A Xanthomonas oryzae pv. oryzae PXO99A]. The December 2014 RCSB PDB [https://pdb.rcsb.org/pdb/static.do?p=education_discussion/molecule_of_the_month/index.html Molecule of the Month] feature on ''TAL Effectors'' by David Goodsell is [https://dx.doi.org/10.2210/rcsb_pdb/mom_2014_12 10.2210/rcsb_pdb/mom_2014_12]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3UGM OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3UGM FirstGlance]. <br> |
| - | </td></tr><tr id=' | + | </td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 3Å</td></tr> |
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ugm FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ugm OCA], [https://pdbe.org/3ugm PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ugm RCSB], [https://www.ebi.ac.uk/pdbsum/3ugm PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ugm ProSAT]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3ugm FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3ugm OCA], [https://pdbe.org/3ugm PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3ugm RCSB], [https://www.ebi.ac.uk/pdbsum/3ugm PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3ugm ProSAT]</span></td></tr> | ||
</table> | </table> | ||
| - | + | == Function == | |
| - | = | + | [https://www.uniprot.org/uniprot/PTHX1_XANOP PTHX1_XANOP] Avirulence protein. Acts as a transcription factor in rice, inducing expression of a number of host genes including SWEET11 (Os8N3, XA13, AC Q6YZF3) in susceptible plants with the Xa13 allele. Plants with the xa13 allele, which has an altered promoter, are resistant to bacterial blight caused by this bacterial strain and do not induce SWEET11. The xa13 allele elicits an atypical hypersensitive response (HR). PthXo1 binds SWEET11 promoter DNA in a sequence-specific manner.<ref>PMID:15553245</ref> <ref>PMID:16798873</ref> <ref>PMID:20345643</ref> <ref>PMID:22223736</ref> |
| - | + | ||
| - | + | ||
| - | The | + | |
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | < | + | |
==See Also== | ==See Also== | ||
| Line 26: | Line 19: | ||
[[Category: RCSB PDB Molecule of the Month]] | [[Category: RCSB PDB Molecule of the Month]] | ||
[[Category: TAL Effectors]] | [[Category: TAL Effectors]] | ||
| - | [[Category: | + | [[Category: Xanthomonas oryzae pv. oryzae PXO99A]] |
| - | [[Category: Bogdanove | + | [[Category: Bogdanove AJ]] |
| - | [[Category: Bradley | + | [[Category: Bradley P]] |
| - | [[Category: Cernadas | + | [[Category: Cernadas RA]] |
| - | [[Category: Mak | + | [[Category: Mak ANS]] |
| - | [[Category: Stoddard | + | [[Category: Stoddard BL]] |
| - | + | ||
| - | + | ||
| - | + | ||
Current revision
Structure of TAL effector PthXo1 bound to its DNA target
| |||||||||||