This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:SaraKathryn Kalkhoff/Sandbox 1
From Proteopedia
| Line 1: | Line 1: | ||
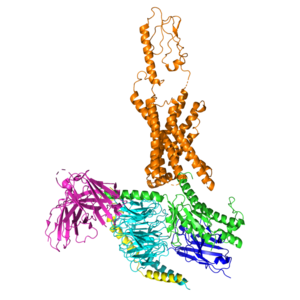
| - | + | <StructureSelection load='7rbt' frame='true' size='350' side='right' caption='Gastric Inhibitory Polypeptide Receptor (GIP-R) bound to Tirzepatide.' scene=''> | |
== Introduction == | == Introduction == | ||
[https://www.sciencedirect.com/topics/medicine-and-dentistry/gastric-inhibitory-polypeptide GIP-R is a receptor that binds glucose within many different cells.] | [https://www.sciencedirect.com/topics/medicine-and-dentistry/gastric-inhibitory-polypeptide GIP-R is a receptor that binds glucose within many different cells.] | ||
Revision as of 14:24, 2 April 2024
<StructureSelection load='7rbt' frame='true' size='350' side='right' caption='Gastric Inhibitory Polypeptide Receptor (GIP-R) bound to Tirzepatide.' scene=>
Contents |
Introduction
GIP-R is a receptor that binds glucose within many different cells.
Function
The GIP receptor helps facilitate movement of glucose within a cell. [1].
Tirzepatide
Tirzepatide has been used as a treatment for Type 2 diabetes. It is used to help treat Type 2 Diabetes, as an agonist to allow insulin to be broken down.
Disease
Diabetes is very bad for you[2].
Relevance
Structural highlights
Active Site
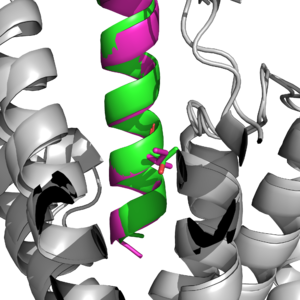
Main binding domains between tirzepatide and the GIP receptor would contain an arginine 190 and glutamine 220 residues to facilitate binding of the ligand. One key difference found was a point mutation at position 7 between an Isoleucine and Threonine [1], which would result in a higher affinity for the tirzepatide molecule binding onto the receptor than the ligand.
References
- ↑ 1.0 1.1 Sun B, Willard FS, Feng D, Alsina-Fernandez J, Chen Q, Vieth M, Ho JD, Showalter AD, Stutsman C, Ding L, Suter TM, Dunbar JD, Carpenter JW, Mohammed FA, Aihara E, Brown RA, Bueno AB, Emmerson PJ, Moyers JS, Kobilka TS, Coghlan MP, Kobilka BK, Sloop KW. Structural determinants of dual incretin receptor agonism by tirzepatide. Proc Natl Acad Sci U S A. 2022 Mar 29;119(13):e2116506119. PMID:35333651 doi:10.1073/pnas.2116506119
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
Student Contributors
SaraKathryn Kalkhoff Camille Gaudet </StructureSection>