This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
User:SaraKathryn Kalkhoff/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 1: | Line 1: | ||
<StructureSection load='7rbt' size='350' frame='true' | <StructureSection load='7rbt' size='350' frame='true' | ||
| - | side='right' caption=' | + | side='right' caption='Glucose-dependent Insulinotropic Polypeptide (GIP) bound to Tirzepatide' scene=’’> |
== Introduction == | == Introduction == | ||
| Line 9: | Line 9: | ||
The GIP receptor helps facilitate movement of glucose within a cell. <ref name='Sun'>PMID:35333651</ref>. It has a natural ligand that is 42 residues and helps kickstart the GIP-R into firing, as a transporter for glucose in and out of the cell. Once the levels become too high, the ligand | The GIP receptor helps facilitate movement of glucose within a cell. <ref name='Sun'>PMID:35333651</ref>. It has a natural ligand that is 42 residues and helps kickstart the GIP-R into firing, as a transporter for glucose in and out of the cell. Once the levels become too high, the ligand | ||
===Tirzepatide=== | ===Tirzepatide=== | ||
| - | Tirzepatide has been used as a treatment for Type 2 diabetes | + | Tirzepatide has been used as a treatment for Type 2 diabetes and <scene name='10/1037492/Just_tirz/1'>Tirzepatide has a total of 39 residues present in its structure.</scene> It is used to help treat Type 2 Diabetes, as an agonist to allow insulin to be broken down. This also works in tandem with GLP-1... It also contains two residues with an AIB sequence, which stands for alpha-amino isobutric acid and aids with preventing degradation of the peptide <ref name='Chavda>PMID: 35807558</ref>. |
== Disease == | == Disease == | ||
| Line 17: | Line 17: | ||
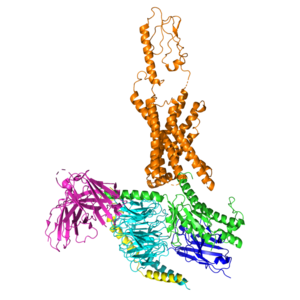
<scene name='10/1037492/Gip_tirz/7'>Main scene of Tirzepatide bound to the GIP receptor. </scene> | <scene name='10/1037492/Gip_tirz/7'>Main scene of Tirzepatide bound to the GIP receptor. </scene> | ||
=== Active Site === | === Active Site === | ||
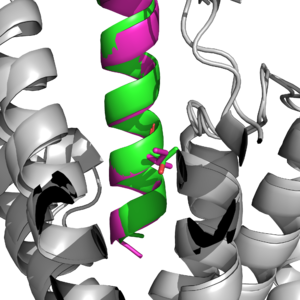
| - | Main binding domains between tirzepatide and the GIP receptor would contain an arginine 190 and glutamine 220 residues to facilitate binding of the ligand. One key difference found was a point mutation at position 7 between an Isoleucine and Threonine <ref name='Sun'>PMID:35333651</ref>, which would result in a higher affinity for the tirzepatide molecule binding onto the receptor than the ligand. | + | Main binding domains between tirzepatide and the GIP receptor would contain an arginine 190 and glutamine 220 residues to facilitate binding of the ligand. The binding is also able to use a tyrosine residue at position 1 that helps guide the ligand into the correct binding spot <author here>.One key difference found was a point mutation at position 7 between an Isoleucine and Threonine <ref name='Sun'>PMID:35333651</ref>, which would result in a higher affinity for the tirzepatide molecule binding onto the receptor than the ligand. |
| - | [[Image:i7_vs_t7.png|300px|left|thumb|Figure 2: Key difference between GIP ligand and Tirzepatide at position 7. Ile7 is in pink (ligand), and Thr7 is in | + | [[Image:i7_vs_t7.png|300px|left|thumb|Figure 2: Key difference between GIP ligand and Tirzepatide at position 7. Ile7 is in pink (ligand), and Thr7 is in aqua(tirzepatide).]] |
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 13:20, 9 April 2024
| |||||||||||