This old version of Proteopedia is provided for student assignments while the new version is undergoing repairs. Content and edits done in this old version of Proteopedia after March 1, 2026 will eventually be lost when it is retired in about June of 2026.
Apply for new accounts at the new Proteopedia. Your logins will work in both the old and new versions.
Andrew Helmerich Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 23: | Line 23: | ||
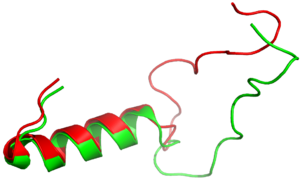
=== Amidated C-Terminus=== | === Amidated C-Terminus=== | ||
| - | <scene name='10/1038819/Amidated_c_term/ | + | <scene name='10/1038819/Amidated_c_term/9'>Amidated C-Terminus</scene> |
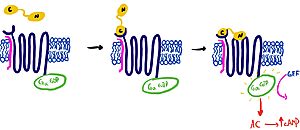
===G-Alpha-TMD Interaction=== | ===G-Alpha-TMD Interaction=== | ||
Revision as of 13:54, 18 April 2024
The Amylin Receptor(AMYR)
| |||||||||||
References
- ↑ Ransey E, Paredes E, Dey SK, Das SR, Heroux A, Macbeth MR. Crystal structure of the Entamoeba histolytica RNA lariat debranching enzyme EhDbr1 reveals a catalytic Zn(2+) /Mn(2+) heterobinucleation. FEBS Lett. 2017 Jul;591(13):2003-2010. doi: 10.1002/1873-3468.12677. Epub 2017, Jun 14. PMID:28504306 doi:http://dx.doi.org/10.1002/1873-3468.12677
- ↑ Cao J, Belousoff MJ, Liang YL, Johnson RM, Josephs TM, Fletcher MM, Christopoulos A, Hay DL, Danev R, Wootten D, Sexton PM. A structural basis for amylin receptor phenotype. Science. 2022 Mar 25;375(6587):eabm9609. PMID:35324283 doi:10.1126/science.abm9609
Student Contributors
- Ty Vander Eide
- Andrew Helmerich
- Ben Whiteside