We apologize for Proteopedia being slow to respond. For the past two years, a new implementation of Proteopedia has been being built. Soon, it will replace this 18-year old system. All existing content will be moved to the new system at a date that will be announced here.
User:Mandy Bechman/Sandbox 1
From Proteopedia
(Difference between revisions)
| Line 14: | Line 14: | ||
== Function == | == Function == | ||
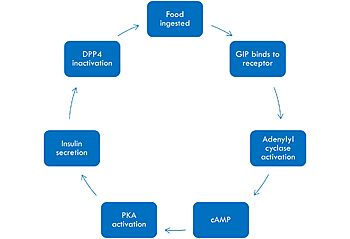
| - | [[Image:GIPmech.jpg|350 px|left|thumb|Mechanism]] | + | [[Image:GIPmech.jpg|350 px|left|thumb|Mechanism of GIP activation and deactivation.]] |
| - | The mechanism of GIP begins with the ingestion of food. This stimulates GIP to bind to its receptor. Once bound, the activated G alpha protein moves laterally in the membrane in order to activate its target, adenylyl cyclase. Once adenylyl cyclase is activated, it catalyzes the formation of cyclic AMP (cAMP). This molecule activates Protein Kinase A (PKA), which signals the secretin of insulin. After a few minutes of active signaling, this | + | The mechanism of GIP begins with the ingestion of food. This stimulates GIP to bind to its receptor. Once bound, the activated G alpha protein moves laterally in the membrane in order to activate its target, adenylyl cyclase. Once adenylyl cyclase is activated, it catalyzes the formation of cyclic AMP (cAMP). This molecule activates Protein Kinase A (PKA), which signals the secretin of insulin. After a few minutes of active signaling, this hormone is recognized and inactivated by a peptidase called DPP-4 (dipeptidyl peptidase-4) by cleaving the first two amino acids. |
Revision as of 14:20, 18 April 2024
H. sapiens Glucose-dependent Insulinotropic Polypeptide
| |||||||||||
References
- ↑ 1.0 1.1 Mayendraraj A, Rosenkilde MM, Gasbjerg LS. GLP-1 and GIP receptor signaling in beta cells interactions and co-stimulation. Peptides. 2022 May;151:170749. PMID:35065096 doi:10.1016/j.peptides.2022.170749
- ↑ 2.0 2.1 Seino Y, Fukushima M, Yabe D. GIP and GLP-1, the two incretin hormones: Similarities and differences. J Diabetes Investig. 2010 Apr 22;1(1-2):8-23. PMID:24843404 doi:10.1111/j.2040-1124.2010.00022.x
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644